8ISO
 
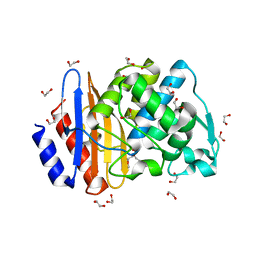 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | 分子名称: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | 著者 | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | 登録日 | 2023-03-21 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISP
 
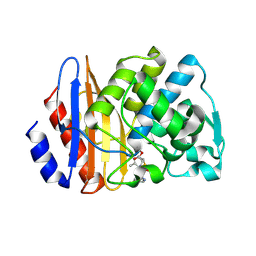 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | 分子名称: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | 著者 | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | 登録日 | 2023-03-21 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
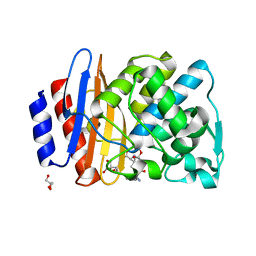 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | 著者 | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | 登録日 | 2023-03-21 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISR
 
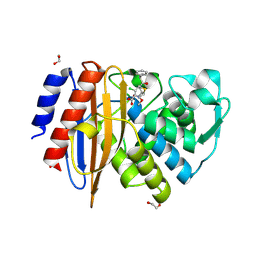 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | 分子名称: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | 著者 | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | 登録日 | 2023-03-21 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
4GQV
 
 | | Crystal structure of CBS-pair protein, CBSX1 from Arabidopsis thaliana | | 分子名称: | CBS domain-containing protein CBSX1, chloroplastic | | 著者 | Jeong, B.-C, Park, S.H, Yoo, K.S, Shin, J.S, Song, H.K. | | 登録日 | 2012-08-24 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.392 Å) | | 主引用文献 | Crystal structure of the single cystathionine beta-synthase domain-containing protein CBSX1 from Arabidopsis thaliana
Biochem.Biophys.Res.Commun., 430, 2013
|
|
4GQW
 
 | |
3SL7
 
 | | Crystal structure of CBS-pair protein, CBSX2 from Arabidopsis thaliana | | 分子名称: | ACETATE ION, CBS domain-containing protein CBSX2, GLYCEROL | | 著者 | Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2011-06-24 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.905 Å) | | 主引用文献 | Single cystathionine beta-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis
Plant Cell, 23, 2011
|
|
7K36
 
 | | Cryo-EM structure of STRIPAK complex | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, MANGANESE (II) ION, MOB-like protein phocein, ... | | 著者 | Jeong, B.-C, Bai, X.C. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of the Hippo signaling integrator human STRIPAK.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4GQY
 
 | | Crystal structure of CBSX2 in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, CBS domain-containing protein CBSX2, chloroplastic | | 著者 | Jeong, B.C, Song, H.K. | | 登録日 | 2012-08-24 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | Change in single cystathionine beta-synthase domain-containing protein from a bent to flat conformation upon adenosine monophosphate binding
J.Struct.Biol., 183, 2013
|
|
5WC2
 
 | | Crystal Structure of ADP-bound human TRIP13 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | 著者 | Jeong, B.-C, Luo, X. | | 登録日 | 2017-06-29 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing.
Nat Commun, 8, 2017
|
|
7VYT
 
 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | 分子名称: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | 著者 | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | 登録日 | 2021-11-15 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
7VYR
 
 | |
7CIN
 
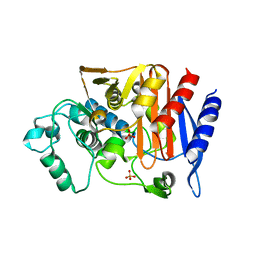 | |
7YTN
 
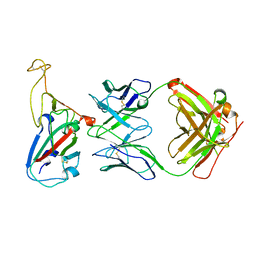 | |
3PO0
 
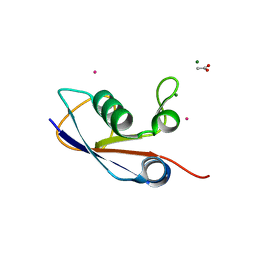 | | Crystal structure of SAMP1 from Haloferax volcanii | | 分子名称: | ACETATE ION, CADMIUM ION, MAGNESIUM ION, ... | | 著者 | Jeong, Y.J, Jeong, B.-C, Song, H.K. | | 登録日 | 2010-11-21 | | 公開日 | 2011-03-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
6V5C
 
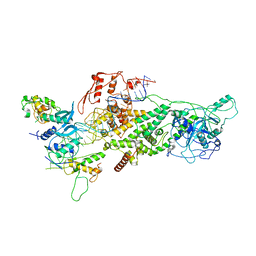 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | 分子名称: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | 著者 | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | 登録日 | 2019-12-04 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
6V5B
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - Active state | | 分子名称: | CALCIUM ION, Microprocessor complex subunit DGCR8, Pri-miR-16-2 (78-MER), ... | | 著者 | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | 登録日 | 2019-12-04 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
3KB5
 
 | |
3NIJ
 
 | | The structure of UBR box (HIAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIT
 
 | | The structure of UBR box (native1) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIH
 
 | | The structure of UBR box (RIAAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NII
 
 | | The structure of UBR box (KIAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIN
 
 | | The structure of UBR box (RLGES) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide RLGES, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIK
 
 | | The structure of UBR box (REAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIL
 
 | | The structure of UBR box (RDAA) | | 分子名称: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
