1HJA
 
 | |
1SGN
 
 | | ASN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with
Streptomyces griseus Protease B at Various pHs
To be Published
|
|
1SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
1AVS
 
 | |
1AVF
 
 | | ACTIVATION INTERMEDIATE 2 OF HUMAN GASTRICSIN FROM HUMAN STOMACH | | Descriptor: | GASTRICSIN, SODIUM ION | | Authors: | Khan, A.R, Cherney, M.M, Tarasova, N.I, James, M.N.G. | | Deposit date: | 1997-09-16 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural characterization of activation 'intermediate 2' on the pathway to human gastricsin.
Nat.Struct.Biol., 4, 1997
|
|
1MBQ
 
 | | Anionic Trypsin from Pacific Chum Salmon | | Descriptor: | BENZAMIDINE, CALCIUM ION, Trypsin | | Authors: | Toyota, E, Ng, K.K.S, Kuninaga, S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Nucleotide Sequence of an Anionic Trypsin from Chum Salmon (Oncorhynchus keta) in Comparison with Atlantic Salmon (Salmo salar) and Bovine Trypsin
J.Mol.Biol., 324, 2002
|
|
1MBM
 
 | | NSP4 proteinase from Equine Arteritis Virus | | Descriptor: | chymotrypsin-like serine protease | | Authors: | Barrette-Ng, I.H, Ng, K.K.-S, Mark, B.L, van Aken, D, Cherney, M.M, Garen, C, Kolodenko, Y, Gorbalenya, A.E, Snijder, E.J, James, M.N.G. | | Deposit date: | 2002-08-03 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Arterivirus nsp4: the smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole
J.Biol.Chem., 277, 2002
|
|
1NHV
 
 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(5-BENZOFURAN-2-YL-THIOPHEN-2-YLMETHYL)-(2,4-DICHLORO-BENZOYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
1NHU
 
 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
1NOU
 
 | | Native human lysosomal beta-hexosaminidase isoform B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
1NP0
 
 | | Human lysosomal beta-hexosaminidase isoform B in complex with intermediate analogue NAG-thiazoline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase subunit beta, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
2SEC
 
 | |
2SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water molecules participate in proteinase-inhibitor interactions: crystal structures of Leu18, Ala18, and Gly18 variants of turkey ovomucoid inhibitor third domain complexed with Streptomyces griseus proteinase B.
Protein Sci., 4, 1995
|
|
2NU1
 
 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I
To be Published
|
|
2NU0
 
 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I
To be Published
|
|
2NR9
 
 | | Crystal structure of GlpG, Rhomboid Peptidase from Haemophilus influenzae | | Descriptor: | (R)-2-(FORMYLOXY)-3-(PHOSPHONOOXY)PROPYL PENTANOATE, 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, Protein glpG homolog | | Authors: | Lemieux, M.J, Fischer, S.J, Cherney, M.M, Bateman, K.S, James, M.N.G. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NU4
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2O2H
 
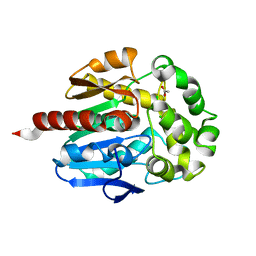 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane | | Descriptor: | 1,2-DICHLOROETHANE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane
TO BE PUBLISHED
|
|
2O2I
 
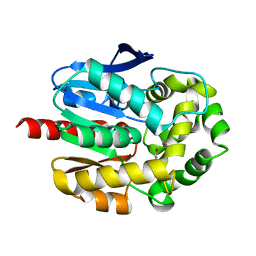 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,3-propandiol | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, Haloalkane dehalogenase 3 | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,3-propandiol
To be Published
|
|
2NU3
 
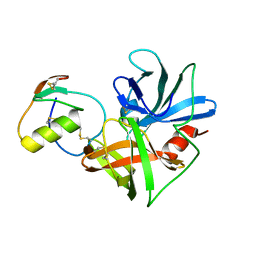 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2NU2
 
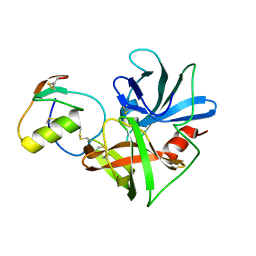 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2Q9H
 
 | | Crystal structure of the C73S mutant of diaminopimelate epimerase | | Descriptor: | ACETIC ACID, Diaminopimelate epimerase, L(+)-TARTARIC ACID | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
1QA7
 
 | | CRYSTAL COMPLEX OF THE 3C PROTEINASE FROM HEPATITIS A VIRUS WITH ITS INHIBITOR AND IMPLICATIONS FOR THE POLYPROTEIN PROCESSING IN HAV | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HAV 3C PROTEINASE, ... | | Authors: | Bergmann, E.M, Cherney, M.M, Mckendrick, J, Vederas, J.C, James, M.N.G. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV.
Virology, 265, 1999
|
|
2Z7A
 
 | | X-ray crystal structure of RV0760c from Mycobacterium tuberculosis at 2.10 Angstrom resolution | | Descriptor: | ACETATE ION, Putative steroid isomerase | | Authors: | Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase.
Biochim.Biophys.Acta, 1784, 2008
|
|
2Z20
 
 | | Crystal structure of LL-Diaminopimelate Aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Cherney, M.M, van Belkum, M.J, Marcus, S.L, Flegel, M.D, Clay, M.D, Deyholos, M.K, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana: a recently discovered enzyme in the biosynthesis of L-lysine by plants and Chlamydia
J.Mol.Biol., 371, 2007
|
|
