2PSG
 
 | |
4SGB
 
 | |
2SGA
 
 | |
3APP
 
 | |
6LBA
 
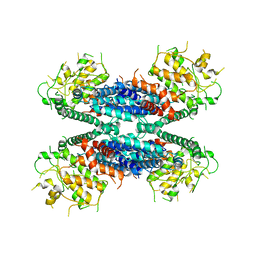 | | Cryo-EM structure of the AtMLKL2 tetramer | | Descriptor: | Protein kinase family protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
2A5A
 
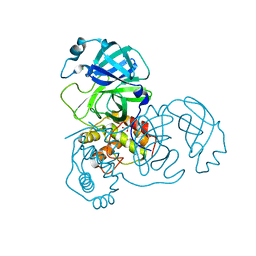 | | Crystal structure of unbound SARS coronavirus main peptidase in the space group C2 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like peptidase, CHLORIDE ION | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
3C9E
 
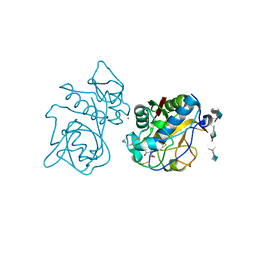 | | Crystal structure of the cathepsin K : chondroitin sulfate complex. | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Kienetz, M, Cherney, M.M, James, M.N.G, Bromme, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex.
J.Mol.Biol., 383, 2008
|
|
1QA7
 
 | | CRYSTAL COMPLEX OF THE 3C PROTEINASE FROM HEPATITIS A VIRUS WITH ITS INHIBITOR AND IMPLICATIONS FOR THE POLYPROTEIN PROCESSING IN HAV | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HAV 3C PROTEINASE, ... | | Authors: | Bergmann, E.M, Cherney, M.M, Mckendrick, J, Vederas, J.C, James, M.N.G. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV.
Virology, 265, 1999
|
|
4MJ2
 
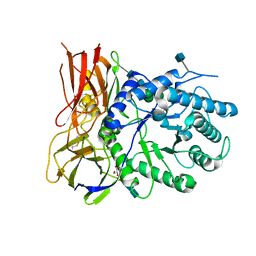 | | Crystal structure of apo-iduronidase in the R3 form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-09-03 | | Release date: | 2013-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
1QRP
 
 | | Human pepsin 3A in complex with a phosphonate inhibitor IVA-VAL-VAL-LEU(P)-(O)PHE-ALA-ALA-OME | | Descriptor: | PEPSIN 3A, methyl N-[(2S)-2-({(S)-hydroxy[(1R)-3-methyl-1-{[N-(3-methylbutanoyl)-L-valyl-L-valyl]amino}butyl]phosphoryl}oxy)-3-phenylpropanoyl]-L-alanyl-L-alaninate | | Authors: | Fujinaga, M, Cherney, M.M, Tarasova, N.I, Bartlett, P.A, Hanson, J.E, James, M.N.G. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural study of the complex between human pepsin and a phosphorus-containing peptidic -transition-state analog.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1HJA
 
 | |
4MJ4
 
 | | Human iduronidase apo structure P21 form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, CHLORIDE ION, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-09-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
1QS8
 
 | | Crystal structure of the P. vivax aspartic proteinase plasmepsin complexed with the inhibitor pepstatin A | | Descriptor: | ACETATE ION, PEPSTATIN A, PLASMEPSIN | | Authors: | Khazanovich Bernstein, N, Cherney, M.M, Yowell, C.A, Dame, J.B, James, M.N.G. | | Deposit date: | 1999-06-25 | | Release date: | 1999-07-07 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation of P. vivax plasmepsin.
J.Mol.Biol., 329, 2003
|
|
2QAA
 
 | |
2QA9
 
 | |
2ATO
 
 | | Crystal structure of Human Cathepsin K in complex with myocrisin | | Descriptor: | (S)-(1,2-DICARBOXYETHYLTHIO)GOLD, Cathepsin K, SULFATE ION | | Authors: | Weidauer, E, Yasuda, Y, Biswal, B.K, Kerr, L.D, Cherney, M.M, Gordon, R.E, James, M.N.G, Bromme, D. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of disease-modifying anti-rheumatic drugs (DMARDs) on the activities of rheumatoid arthritis-associated cathepsins K and S.
Biol.Chem., 388, 2007
|
|
4SGA
 
 | |
2A5I
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in the space group C2 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 1,2-ETHANEDIOL, 3C-like peptidase, ... | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5K
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2Q9H
 
 | | Crystal structure of the C73S mutant of diaminopimelate epimerase | | Descriptor: | ACETIC ACID, Diaminopimelate epimerase, L(+)-TARTARIC ACID | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2A15
 
 | | X-ray Crystal Structure of RV0760 from Mycobacterium Tuberculosis at 1.68 Angstrom Resolution | | Descriptor: | HYPOTHETICAL PROTEIN Rv0760c, NICOTINAMIDE | | Authors: | Garen, C.R, Cherney, M.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase
Biochim.Biophys.Acta, 1784, 2008
|
|
2AQ6
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase complexed with pyridoxal 5'-phosphate at 1.7 a resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRIDOXINE 5'-PHOSPHATE OXIDASE | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Mycobacterium tuberculosispyridoxine 5'-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5TNC
 
 | |
3SGQ
 
 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18,
Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
3SGB
 
 | | STRUCTURE OF THE COMPLEX OF STREPTOMYCES GRISEUS PROTEASE B AND THE THIRD DOMAIN OF THE TURKEY OVOMUCOID INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | PROTEINASE B (SGPB), TURKEY OVOMUCOID INHIBITOR (OMTKY3) | | Authors: | Read, R.J, Fujinaga, M, Sielecki, A.R, James, M.N.G. | | Deposit date: | 1983-01-21 | | Release date: | 1983-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the complex of Streptomyces griseus protease B and the third domain of the turkey ovomucoid inhibitor at 1.8-A resolution.
Biochemistry, 22, 1983
|
|
