1BDK
 
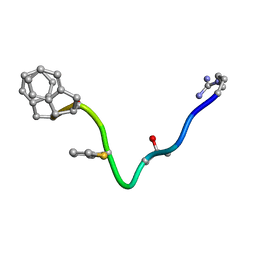 | | AN NMR, CD, MOLECULAR DYNAMICS, AND FLUOROMETRIC STUDY OF THE CONFORMATION OF THE BRADYKININ ANTAGONIST B-9340 IN WATER AND IN AQUEOUS MICELLAR SOLUTIONS | | Descriptor: | bradykinin antagonist B-9340 | | Authors: | Sejbal, J, Kotovych, G, Cann, J.R, Stewart, J.M, Gera, L. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-07 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | An NMR, CD, molecular dynamics, and fluorometric study of the conformation of the bradykinin antagonist B-9340 in water and in aqueous micellar solutions.
J.Med.Chem., 39, 1996
|
|
1BKL
 
 | | SELF-ASSOCIATED APO SRC SH2 DOMAIN | | Descriptor: | PP60 V-SRC TYROSINE KINASE TRANSFORMING PROTEIN | | Authors: | Holland, D.R, Rubin, J.R. | | Deposit date: | 1997-05-02 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel Pp60Src Sh2 Domain Crystal Structures: A 2.0 Angstrom Co-Crystal Structure of a D-Amino Acid Substituted Phosphopeptide Complex and a 2.1 Angstrom Apo Structure Displaying Self-Association
To be Published
|
|
7SFF
 
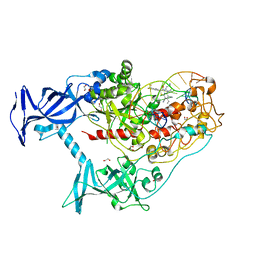 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3852279B | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]sulfanyl}-2-[4-(trifluoromethyl)phenyl]acetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFD
 
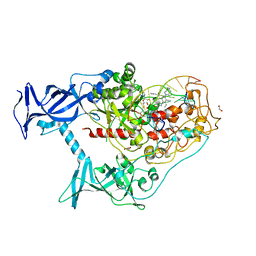 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3543105A | | Descriptor: | (2S)-2-({3,5-dicyano-4-ethyl-6-[4-(2-hydroxyethyl)-1,4-diazepan-1-yl]pyridin-2-yl}sulfanyl)-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFC
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3735967A | | Descriptor: | 1,2-ETHANEDIOL, DNA (12-MER), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFE
 
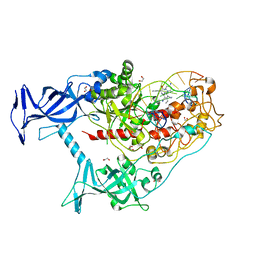 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830334A | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]amino}-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFG
 
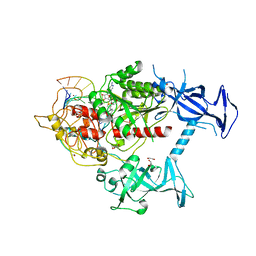 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Cofactor SAM | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 1, DNA Strand 1, ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
2L79
 
 | |
2L77
 
 | |
8SO0
 
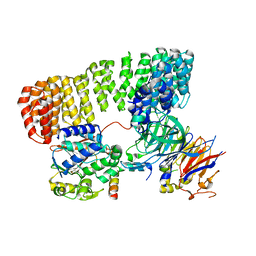 | | Cryo-EM structure of the PP2A:B55-FAM122A complex | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-04-28 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.79961 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
2LEK
 
 | | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris refined with NH RDCs. Northeast Structural Genomics Consortium target RpR325 | | Descriptor: | Putative thiamin biosynthesis ThiS | | Authors: | Ramelot, T.A, Cort, J.R, Lee, H, Wang, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium target RpR325
To be Published
|
|
1C3P
 
 | |
1C3S
 
 | |
1BVX
 
 | | THE 1.8 A STRUCTURE OF GEL GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C, Helliwell, J.R. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7KD1
 
 | |
1D0I
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR COMPLEXED WITH PHOSPHATE IONS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, TYPE II 3-DEHYDROQUINATE HYDRATASE | | Authors: | Roszak, A.W, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1999-09-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and mechanism of the type II dehydroquinase from Streptomyces coelicolor.
Structure, 10, 2002
|
|
8T9U
 
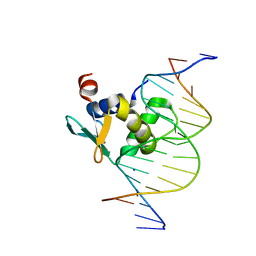 | | Human PU.1 ETS-domain (165-270) in complex with d(AATAAGCGIAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*IP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dissection of integrated readout reveals the structural thermodynamics of DNA selection by transcription factors.
Structure, 32, 2024
|
|
8SMH
 
 | |
1D49
 
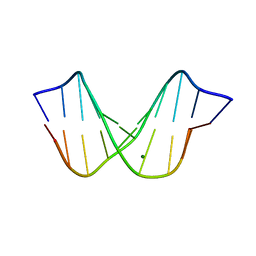 | | THE STRUCTURE OF A B-DNA DECAMER WITH A CENTRAL T-A STEP: C-G-A-T-T-A-A-T-C-G | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*AP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Grzeskowiak, K, Yanagi, K, Dickerson, R.E. | | Deposit date: | 1991-09-17 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G.
J.Mol.Biol., 225, 1992
|
|
8SP1
 
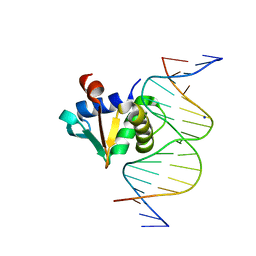 | |
1D43
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE UP | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
8SMJ
 
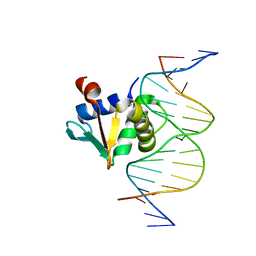 | |
3PPA
 
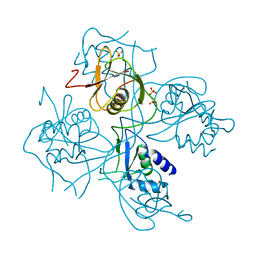 | | Structure of the Dusp-Ubl domains of Usp15 | | Descriptor: | CITRIC ACID, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Walker, J.R, Asinas, A.E, Dong, A, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Dusp-Ubl Domains of the Ubiquitin-Specific Protease 15
To be Published
|
|
2LO8
 
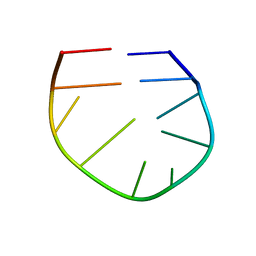 | |
2LO5
 
 | | Structural Basis for Bifunctional Zn(II) Macrocyclic Complex Recognition of Thymine Bulges in DNA, Structure of a Thymine bulge | | Descriptor: | DNA (5'-D(*GP*GP*CP*CP*GP*CP*AP*GP*TP*GP*CP*C)-3') | | Authors: | Morrow, J.R, Fountain, M.A, del Mundo, I.A, Siter, K.E. | | Deposit date: | 2012-01-17 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Bifunctional Zinc(II) Macrocyclic Complex Recognition of Thymine Bulges in DNA.
Inorg.Chem., 51, 2012
|
|
