4I91
 
 | | Crystal Structure of Cytochrome P450 2B6 (Y226H/K262R) in complex with alpha-Pinene. | | 分子名称: | (+)-alpha-Pinene, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | 著者 | Shah, M.B, Stout, C.D, Halpert, J.R. | | 登録日 | 2012-12-04 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Thermodynamic Basis of (+)-alpha-Pinene Binding to Human Cytochrome P450 2B6.
J.Am.Chem.Soc., 135, 2013
|
|
5EH2
 
 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence III | | 分子名称: | DNA (5'-D(*AP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | 著者 | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | 登録日 | 2015-10-27 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EI9
 
 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence I | | 分子名称: | DNA (5'-D(*AP*TP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*GP*A)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | 著者 | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | 登録日 | 2015-10-29 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.921 Å) | | 主引用文献 | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
3ZVE
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 84 | | 分子名称: | 3C PROTEASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
5A42
 
 | | Cryo-EM single particle 3D reconstruction of the native conformation of E. coli alpha-2-macroglobulin (ECAM) | | 分子名称: | UNCHARACTERIZED LIPOPROTEIN YFHM | | 著者 | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, F.X. | | 登録日 | 2015-06-04 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Structural and Functional Insights Into Escherichia Coli Alpha2- Macroglobulin Endopeptidase Snap-Trap Inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5EGB
 
 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence II | | 分子名称: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | 著者 | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | 登録日 | 2015-10-26 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.977 Å) | | 主引用文献 | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
3ZV9
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 74 | | 分子名称: | 3C PROTEASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZYD
 
 | | Crystal structure of 3C protease of coxsackievirus B3 | | 分子名称: | 3C PROTEINASE, GLYCEROL | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-22 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Peptidic Ab-Nonsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3MVD
 
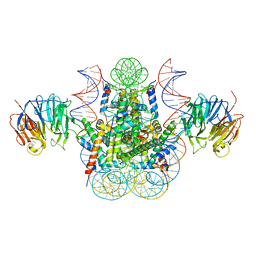 | | Crystal structure of the chromatin factor RCC1 in complex with the nucleosome core particle | | 分子名称: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Makde, R.D, England, J.R, Yennawar, H.P, Tan, S. | | 登録日 | 2010-05-04 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of RCC1 chromatin factor bound to the nucleosome core particle.
Nature, 467, 2010
|
|
8EHW
 
 | |
3ZVC
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 82 | | 分子名称: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(3-TERT-BUTOXY-3-OXOPROPYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
4IQN
 
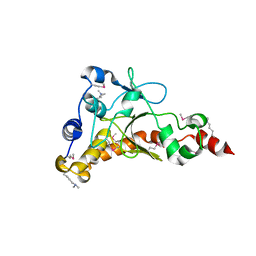 | | Crystal structure of uncharacterized protein from Salmonella enterica subsp. enterica serovar typhimurium str. 14028s | | 分子名称: | DI(HYDROXYETHYL)ETHER, Putative cytoplasmic protein, TETRAETHYLENE GLYCOL | | 著者 | Chang, C, Hatzos-Skintges, C, Adkins, J.N, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | 登録日 | 2013-01-11 | | 公開日 | 2013-01-23 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of uncharacterized protein from salmonella enterica subsp. enterica serovar typhimurium str. 14028s
To be Published
|
|
3MTN
 
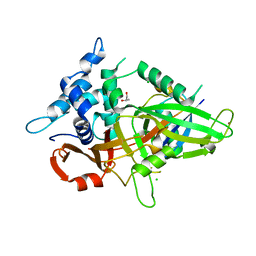 | | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor | | 分子名称: | CHLORIDE ION, GLYCEROL, UBIQUITIN VARIANT UBV.21.4, ... | | 著者 | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2010-04-30 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3ZVG
 
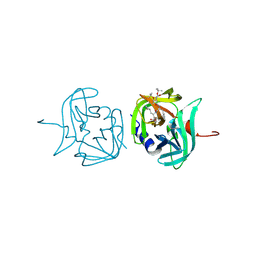 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 98 | | 分子名称: | 3C PROTEASE, N-(tert-butoxycarbonyl)-O-tert-butyl-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZDA
 
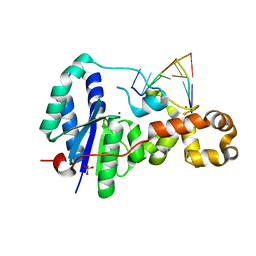 | | Structure of E. coli ExoIX in complex with a fragment of the Flap1 DNA oligonucleotide, potassium and magnesium | | 分子名称: | 5'-D(*AP*AP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP)-3', MAGNESIUM ION, ... | | 著者 | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2012-11-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
5CT9
 
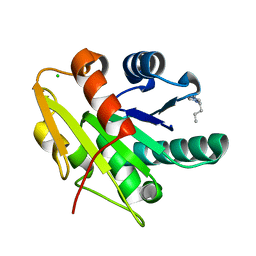 | | G158E/K44E/R57E/Y49E Bacillus subtilis lipase A with 5% [BMIM][Cl] | | 分子名称: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Esterase | | 著者 | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | 登録日 | 2015-07-23 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5CRI
 
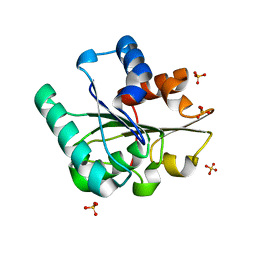 | | Wild-type Bacillus subtilis lipase A with 0% [BMIM][Cl] | | 分子名称: | Esterase, SULFATE ION | | 著者 | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | 登録日 | 2015-07-22 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
4FZ5
 
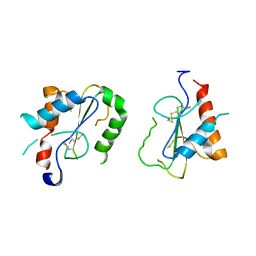 | | Crystal Structure of Human TIRAP TIR-domain | | 分子名称: | Toll/interleukin-1 receptor domain-containing adapter protein | | 著者 | Woo, J.R, Kim, S, Shoelson, S.E, Park, S. | | 登録日 | 2012-07-06 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | X-ray Crystallographic Structure of TIR-Domain from the Human TIR-Domain Containing Adaptor Protein/MyD88 Adaptor-Like Protein (TIRAP/MAL)
Bull.Korean Chem.Soc., 33, 2013
|
|
5CT6
 
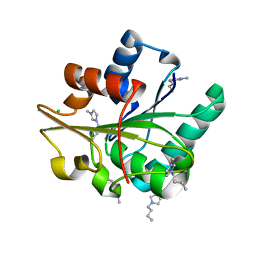 | | Wild-type Bacillus subtilis lipase A with 20% [BMIM][Cl] | | 分子名称: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Lipase EstA | | 著者 | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | 登録日 | 2015-07-23 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5CT8
 
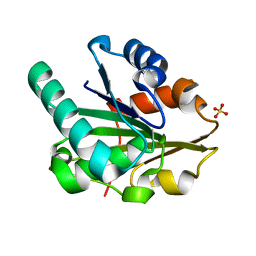 | | G158E/K44E/R57E/Y49E Bacillus subtilis lipase A with 0% [BMIM][Cl] | | 分子名称: | Quadruple mutant lipase A, SULFATE ION | | 著者 | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | 登録日 | 2015-07-23 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
3ZYE
 
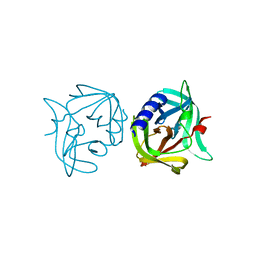 | | Crystal structure of 3C protease mutant (T68A) of coxsackievirus B3 | | 分子名称: | 3C PROTEINASE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-22 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
4G2K
 
 | | Crystal structure of the Marburg Virus GP2 ectodomain in its post-fusion conformation | | 分子名称: | CHLORIDE ION, GLYCEROL, General control protein GCN4, ... | | 著者 | Malashkevich, V.N, Koellhoffer, J.F, Harrison, J.S, Toro, R, Bhosle, R.C, Chandran, K, Lai, J.R, Almo, S.C. | | 登録日 | 2012-07-12 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Marburg Virus GP2 Core Domain in Its Postfusion Conformation.
Biochemistry, 51, 2012
|
|
3MZL
 
 | |
5CT4
 
 | | Wild-type Bacillus subtilis lipase A with 5% [BMIM][Cl] | | 分子名称: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Esterase, ... | | 著者 | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | 登録日 | 2015-07-23 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
3ZVA
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 75 | | 分子名称: | 3C PROTEASE, ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
