7N5W
 
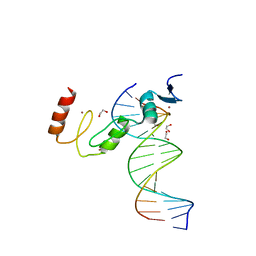 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 23) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand I, DNA Strand II, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5V
 
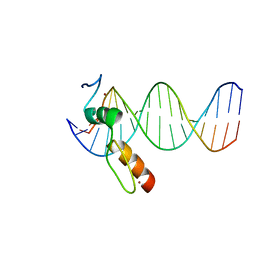 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
2I7A
 
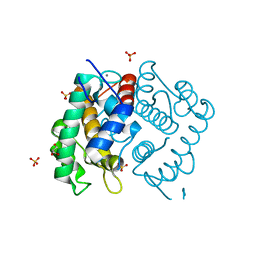 | | Domain IV of Human Calpain 13 | | Descriptor: | CALCIUM ION, Calpain 13, GLYCEROL, ... | | Authors: | Walker, J.R, Ng, K, Davis, T.L, Ravulapalli, R, Butler-cole, C, Finerty Jr, P.J, Newman, E.M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Calpain 13
To be Published
|
|
2JKP
 
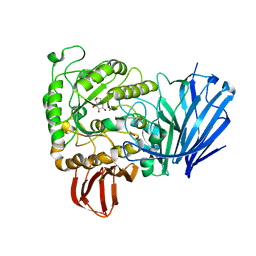 | | Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with castanospermine | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GLUCOSIDASE (ALPHA-GLUCOSIDASE SUSB), CALCIUM ION, ... | | Authors: | Gloster, T.M, Turkenburg, J.P, Potts, J.R, Henrissat, B, Davies, G.J. | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Divergence of Catalytic Mechanism within a Glycosidase Family Provides Insight Into Evolution of Carbohydrate Metabolism by Human Gut Flora.
Chem.Biol., 15, 2008
|
|
2JN8
 
 | | Solution NMR structure of Q8ZRJ2 from Salmonella typhimurium. Northeast Structural Genomics target StR65. | | Descriptor: | Putative cytoplasmic protein | | Authors: | Aramini, J.M, Cort, J.R, Ho, C.K, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-29 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Q8ZRJ2 from Salmonella typhimurium. Northeast Structural Genomics target StR65.
To be Published
|
|
2JNE
 
 | | NMR structure of E.coli YfgJ modelled with two Zn+2 bound. Northeast Structural Genomics Consortium Target ER317. | | Descriptor: | Hypothetical protein yfgJ, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Lukin, J.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of E.coli YfgJ bound to two Zn+2.
To be Published
|
|
2JO6
 
 | | NMR structure of the E.coli protein NirD, Northeast Structural Genomics target ET100 | | Descriptor: | Nitrite reductase [NAD(P)H] small subunit | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Guido, V, Lukin, J.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of E.coli NirD.
To be Published
|
|
2JR2
 
 | | Solution NMR structure of homodimer CPS_2611 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target CsR4. | | Descriptor: | UPF0352 protein CPS_2611 | | Authors: | Ramelot, T.A, Cort, J.R, Wang, H, Nwosu, C, Cunningham, K, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-19 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of homodimer CPS_2611 from Colwellia psychrerythraea.
To be Published
|
|
2JKA
 
 | | Native structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GLUCOSIDASE (ALPHA-GLUCOSIDASE SUSB), CALCIUM ION | | Authors: | Gloster, T.M, Turkenburg, J.P, Potts, J.R, Henrissat, B, Davies, G.J. | | Deposit date: | 2008-08-23 | | Release date: | 2008-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Divergence of Catalytic Mechanism within a Glycosidase Family Provides Insight Into Evolution of Carbohydrate Metabolism by Human Gut Flora.
Chem.Biol., 15, 2008
|
|
2KMM
 
 | | Solution NMR structure of the TGS domain of PG1808 from Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target PgR122A (418-481) | | Descriptor: | Guanosine-3',5'-bis(Diphosphate) 3'-pyrophosphohydrolase | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the TGS domain of PG1808 from
Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target
PgR122A (418-481)
To be Published
|
|
7NJZ
 
 | | X-ray crystallography study of RoAb13 which binds to PIYDIN, a part of the CCR5 N terminal domain | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Helliwell, J.R, Chayen, N, Saridakis, E, Govada, L. | | Deposit date: | 2021-02-17 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
2HE9
 
 | | Structure of the peptidylprolyl isomerase domain of the human NK-tumour recognition protein | | Descriptor: | NK-tumor recognition protein, SULFATE ION | | Authors: | Walker, J.R, Davis, T, Newman, E.M, MacKenzie, F, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2HEQ
 
 | | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399. | | Descriptor: | YorP protein | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.M, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-21 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399.
To be Published
|
|
7OD1
 
 | |
7NT8
 
 | | Influenza virus H3N2 nucleoprotein - R416A mutant | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Nucleoprotein | | Authors: | Keown, J.R, Knight, M.L, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure of an H3N2 influenza virus nucleoprotein.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2KJ7
 
 | | Three-Dimensional NMR Structure of Rat Islet Amyloid Polypeptide in DPC micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Nanga, R, Brender, J.R, Xu, J, Hartman, K, Subramanian, V, Ramamoorthy, A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy.
J.Am.Chem.Soc., 131, 2009
|
|
2KKO
 
 | | Solution NMR structure of the homodimeric winged helix-turn-helix DNA-binding domain (fragment 1-100) Mb0332 from Mycobacterium bovis, a possible ArsR-family transcriptional regulator. Northeast Structural Genomics Consortium Target MbR242E. | | Descriptor: | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (POSSIBLY ARSR-FAMILY) | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the homodimeric winged helix-turn-helix DNA-binding domain (fragment 1-100) Mb0332 from Mycobacterium bovis, a possible ArsR-family transcriptional regulator. Northeast Structural Genomics Consortium Target MbR242E.
To be Published
|
|
7MF3
 
 | | Structure of the autoinhibited state of smooth muscle myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light polypeptide 6, ... | | Authors: | Heissler, S.M, Arora, A.S, Billington, N, Sellers, J.R, Chinthalapudi, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the autoinhibited state of myosin-2.
Sci Adv, 7, 2021
|
|
2KF2
 
 | | Solution NMR structure of of Streptomyces coelicolor polyketide cyclase SCO5315. Northeast Structural Genomics Consortium target RR365 | | Descriptor: | Putative polyketide cyclase | | Authors: | Cort, J.R, Ramelot, T.A, Ding, K, Wang, H, Jiang, M, Maglaqui, M, Xiao, R, Nair, R, Baran, M.C, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-11 | | Release date: | 2009-05-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Streptomyces coelicolor polyketide cyclase SCO5315
To be Published
|
|
2J98
 
 | | Human coronavirus 229E non structural protein 9 cys69ala mutant (Nsp9) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, REPLICASE POLYPROTEIN 1AB | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
2J97
 
 | | Human coronavirus 229E non structural protein 9 (Nsp9) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, REPLICASE POLYPROTEIN 1AB, SULFATE ION | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
2IRW
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Ether Inhibitor | | Descriptor: | (1S,3R,4S,5S,7S)-4-{[2-(4-METHOXYPHENOXY)-2-METHYLPROPANOYL]AMINO}ADAMANTANE-1-CARBOXAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Longenecker, K.L, Patel, J.R, Russell, J, Qin, W. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of adamantane ethers as inhibitors of 11beta-HSD-1: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2H5M
 
 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus complexed with acetyl-CoA. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family | | Authors: | Cort, J.R, Ramelot, T.A, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins
J.Struct.Funct.Genom., 9, 2008
|
|
7OI1
 
 | |
2H01
 
 | | PY00414- Plasmodium yoelii thioredoxin peroxidase I | | Descriptor: | 2-CYS PEROXIREDOXIN | | Authors: | Artz, J, Qiu, W, Min, J.R, Dong, A, Lew, J, Melone, M, Alam, Z, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-12 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of PY00414 - a Plasmodium yoelii thioredoxin peroxidase I
To be Published
|
|
