6UKH
 
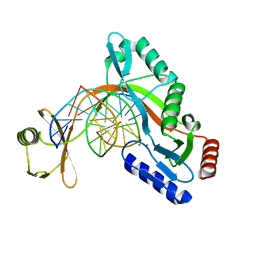 | | HhaI endonuclease in Complex with DNA in space group P41212 (pH 6.0) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*GP*CP*GP*CP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*GP*CP*GP*CP*TP*TP*GP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2019-10-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure of HhaI endonuclease with cognate DNA at an atomic resolution of 1.0 angstrom.
Nucleic Acids Res., 48, 2020
|
|
7KEI
 
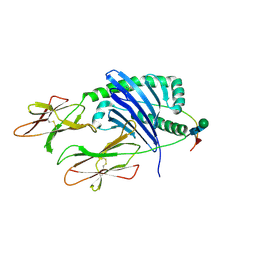 | | DQA1*01:02/DQB1*06:02 in complex with a hemagglutinin peptide from the H1N1 pandemic flu virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA peptide from 2009 H1N1 pandemic flu virus., ... | | Authors: | Birtley, J.R, Stern, L.J, Mellins, E.D, Jiang, W. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of DQA1*01:02/DQB1*06:02 in complex with a flu peptide.
To Be Published
|
|
4UB8
 
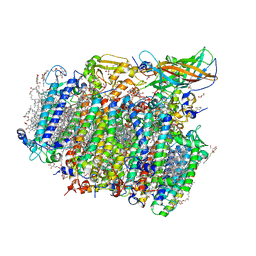 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
1RFB
 
 | |
6V62
 
 | | SETD3 double mutant (N255F/W273A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | An engineered variant of SETD3 methyltransferase alters target specificity from histidine to lysine methylation.
J.Biol.Chem., 295, 2020
|
|
6UTE
 
 | | Crystal structure of Z032 Fab in complex with WNV EDIII | | Descriptor: | Envelope domain III, GLYCEROL, Z032 Fab heavy chain, ... | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UYA
 
 | | Crystal structure of Compound 19 bound to IRAK4 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{2-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-6-(morpholin-4-yl)-1-oxo-2,3-dihydro-1H-isoindol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Kiefer, J.R, Bryan, M.C, Lupardus, P.J, Zarrin, A.A, Rajapaksa, N.S, Gobbi, A, Drobnick, J, Kolesnikov, A, Liang, J, Do, S. | | Deposit date: | 2019-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Potent Benzolactam IRAK4 Inhibitors with Robust in Vivo Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
1ZC4
 
 | | Crystal structure of the Ral-binding domain of Exo84 in complex with the active RalA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Ral-A, ... | | Authors: | Jin, R, Junutula, J.R, Matern, H.T, Ervin, K.E, Scheller, R.H, Brunger, A.T. | | Deposit date: | 2005-04-10 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase.
Embo J., 24, 2005
|
|
4UB6
 
 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
1YWU
 
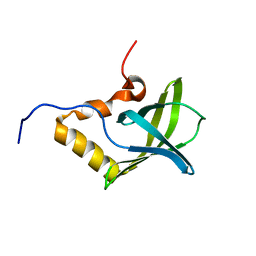 | | Solution NMR structure of Pseudomonas Aeruginosa protein PA4608. Northeast Structural Genomics target PaT7 | | Descriptor: | hypothetical protein PA4608 | | Authors: | Ramelot, T.A, Yee, A.A, Cort, J.R, Semesi, A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein.
Proteins, 66, 2007
|
|
4UQ6
 
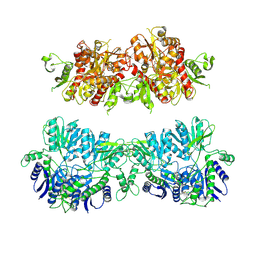 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
1YWW
 
 | |
7L4X
 
 | |
7L4Y
 
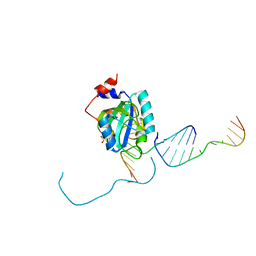 | |
1ZIV
 
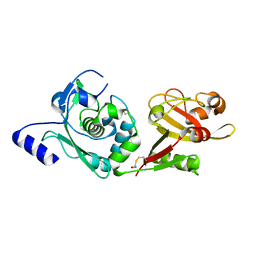 | | Catalytic Domain of Human Calpain-9 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain 9 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Dong, A, Choe, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition.
J.Mol.Biol., 366, 2007
|
|
6WAP
 
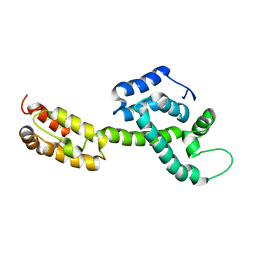 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7L4V
 
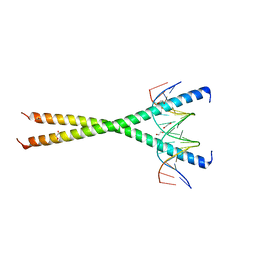 | |
6WE8
 
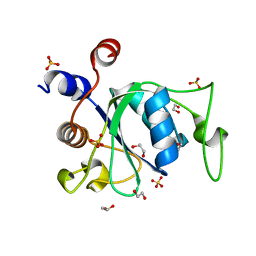 | | YTH domain of human YTHDC1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
6WJS
 
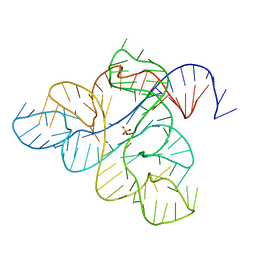 | |
2C4G
 
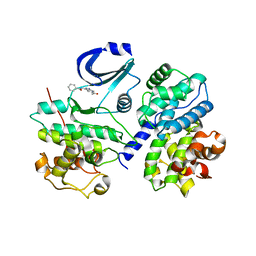 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-533514 | | Descriptor: | (3Z)-5-ACETYL-3-(BENZOYLIMINO)-3,6-DIHYDROPYRROLO[3,4-C]PYRAZOL-5-IUM, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Fancelli, D, Vulpetti, A, Amici, R, Villa, M, Pittala, V, Ciomei, M, Mercurio, C, Bischoff, J.R, Roletto, F, Varasi, M, Brasca, M.G. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 3-Amino-1,4,5,6-Tetrahydropyrrolo[3,4-C]Pyrazoles: A New Class of Cdk2 Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2BCE
 
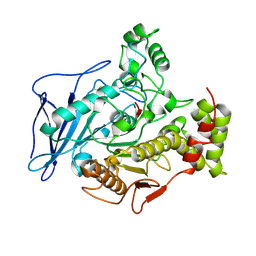 | | CHOLESTEROL ESTERASE FROM BOS TAURUS | | Descriptor: | CHOLESTEROL ESTERASE | | Authors: | Chen, J.C.-H, Miercke, L.J.W, Krucinski, J, Starr, J.R, Saenz, G, Wang, X, Spilburg, C.A, Lange, L.G, Ellsworth, J.L, Stroud, R.M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic cholesterol esterase at 1.6 A: novel structural features involved in lipase activation.
Biochemistry, 37, 1998
|
|
1PXX
 
 | | CRYSTAL STRUCTURE OF DICLOFENAC BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2 | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Rowlinson, S.W, Prusakiewicz, J.J, Pawlitz, J.L, Kozak, K.R, Kalgutkar, A.S, Stallings, W.C, Marnett, L.J, Kurumbail, R.G. | | Deposit date: | 2003-07-07 | | Release date: | 2003-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Mechanism of Cyclooxygenase-2 Inhibition Involving Interactions with Ser-530 and Tyr-385.
J.Biol.Chem., 278, 2003
|
|
2C4C
 
 | | Crystal structure of the NADPH-treated monooxygenase domain of MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-10-18 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1PO5
 
 | | Structure of mammalian cytochrome P450 2B4 | | Descriptor: | Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, He, Y.A, Wester, M.R, White, M.A, Chin, C.C, Halpert, J.R, Johnson, E.F, Stout, C.D. | | Deposit date: | 2003-06-13 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An open conformation of mammalian cytochrome P450 2B4 at 1.6 A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7KRA
 
 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to Fab-DH4 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
