4IHK
 
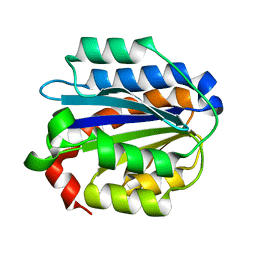 | | Crystal structure of the Collagen VI alpha3 N5 domain R1061Q | | Descriptor: | Collagen alpha3(VI) | | Authors: | Mikolajek, H, Becker, A.K.A, Paulsson, M, Wagener, R, Werner, J.M. | | Deposit date: | 2012-12-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A structure of a collagen VI VWA domain displays N and C termini at opposite sides of the protein
Structure, 22, 2014
|
|
2WYV
 
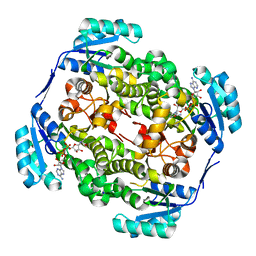 | | High resolution structure of Thermus thermophilus enoyl-acyl carrier protein reductase NAD-form | | Descriptor: | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Otero, J.M, Noel, A.J, Guardado-Calvo, P, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2009-11-20 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | High-resolution structures of Thermus thermophilus enoyl-acyl carrier protein reductase in the apo form, in complex with NAD+ and in complex with NAD+ and triclosan.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
2WV9
 
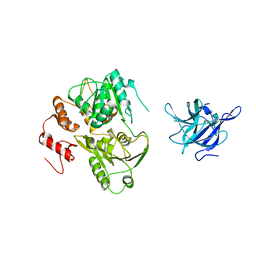 | | Crystal Structure of the NS3 protease-helicase from Murray Valley encephalitis virus | | Descriptor: | FLAVIVIRIN PROTEASE NS2B REGULATORY SUBUNIT, FLAVIVIRIN PROTEASE NS3 CATALYTIC SUBUNIT | | Authors: | Assenberg, R, Mastrangelo, E, Walter, T.S, Verma, A, Milani, M, Owens, R.J, Stuart, D.I, Grimes, J.M, Mancini, E.J. | | Deposit date: | 2009-10-15 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Novel Conformational State of the Flavivirus Ns3 Protein: Implications for Polyprotein Processing and Viral Replication.
J.Virol., 83, 2009
|
|
2WYU
 
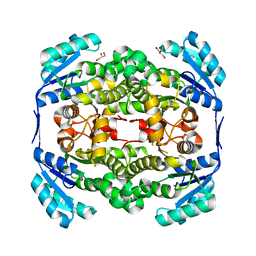 | | High resolution structure of Thermus thermophilus enoyl-acyl carrier protein reductase apo-form | | Descriptor: | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE, GLYCEROL, SODIUM ION | | Authors: | Otero, J.M, Noel, A.J, Guardado-Calvo, P, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2009-11-20 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structures of Thermus thermophilus enoyl-acyl carrier protein reductase in the apo form, in complex with NAD+ and in complex with NAD+ and triclosan.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
2WY8
 
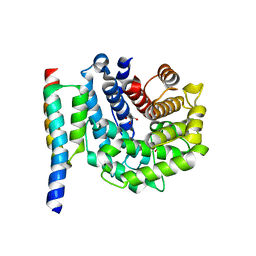 | | Staphylococcus aureus complement subversion protein Sbi-IV in complex with complement fragment C3d | | Descriptor: | COMPLEMENT C3D FRAGMENT, GLYCEROL, IGG-BINDING PROTEIN | | Authors: | Clark, E.A, Crennell, S, Upadhyay, A, Mackay, J.D, Bagby, S, van den Elsen, J.M. | | Deposit date: | 2009-11-13 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Structural Basis for Staphylococcal Complement Subversion: X-Ray Structure of the Complement- Binding Domain of Staphylococcus Aureus Protein Sbi in Complex with Ligand C3D.
Mol.Immunol., 48, 2011
|
|
2X1E
 
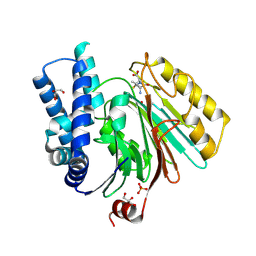 | | The crystal structure of mature acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum in complex 6- aminopenicillanic acid | | Descriptor: | (2S,5R,6R)-6-AMINO-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ACYL-COENZYME, CHLORIDE ION, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
6GKC
 
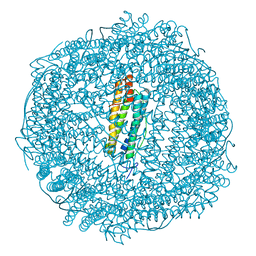 | | 2 minute Fe2+ soak structure of SynFtn | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2018-05-18 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Reaction of O2with a diiron protein generates a mixed-valent Fe2+/Fe3+center and peroxide.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2WSJ
 
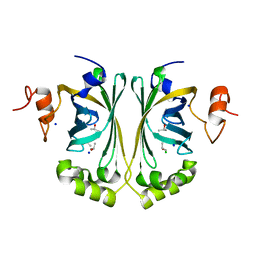 | | Crystal Structure of single point mutant Glu71Ser p-coumaric Acid Decarboxylase | | Descriptor: | BARIUM ION, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Rodriguez, H, Angulo, I, De Las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2009-09-07 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
4HV3
 
 | | Structure of Ricin A chain bound with N-(N-(pterin-7-yl)carbonyl-L-serinyl)-L-tryptophan | | Descriptor: | (2S)-2-[[(2S)-2-[(2-azanyl-4-oxidanylidene-1H-pteridin-7-yl)carbonylamino]-3-oxidanyl-propanoyl]amino]-3-(1H-indol-3-yl)propanoic acid, MALONIC ACID, Ricin, ... | | Authors: | Robertus, J.D, Manzano, L.A, Jasheway, K.R, Monzingo, A.F, Saito, R, Pruet, J.M, Wiget, P.A, Anslyn, E.V. | | Deposit date: | 2012-11-05 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Peptide-conjugated pterins as inhibitors of ricin toxin A.
J.Med.Chem., 56, 2013
|
|
2WO6
 
 | | Human Dual-Specificity Tyrosine-Phosphorylation-Regulated Kinase 1A in complex with a consensus substrate peptide | | Descriptor: | ARTIFICIAL CONSENSUS SEQUENCE, CHLORIDE ION, DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION- REGULATED KINASE 1A, ... | | Authors: | Roos, A.K, Soundararajan, M, Elkins, J.M, Fedorov, O, Eswaran, J, Phillips, C, Pike, A.C.W, Ugochukwu, E, Muniz, J.R.C, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Down Syndrome Kinases, Dyrks, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
6GL8
 
 | | Crystal structure of Bcl-2 in complex with the novel orally active inhibitor S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Casara, P, Davidson, J, Claperon, A, Le Toumelin-Braizat, G, Vogler, M, Bruno, A, Chanrion, M, Lysiak-Auvity, G, Le Diguarher, T, Starck, J.B, Chen, I, Whitehead, N, Graham, C, Matassova, N, Dokurno, P, Pedder, C, Wang, Y, Qiu, S, Girard, A.M, Schneider, E, Grave, F, Studeny, A, Guasconi, G, Rocchetti, F, Maiga, S, Henlin, J.M, Colland, F, Kraus-Berthier, L, Le Gouill, S, Dyer, M.J.S, Hubbard, R, Wood, M, Amiot, M, Cohen, G.M, Hickman, J.A, Morris, E, Murray, J, Geneste, O. | | Deposit date: | 2018-05-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S55746 is a novel orally active BCL-2 selective and potent inhibitor that impairs hematological tumor growth.
Oncotarget, 9, 2018
|
|
2X18
 
 | | The crystal structure of the PH domain of human AKT3 protein kinase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RAC-GAMMA SERINE/THREONINE-PROTEIN KINASE | | Authors: | Vollmar, M, Wang, J, Zhang, Y, Elkins, J.M, Burgess-Brown, N, Chaikuad, A, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2009-12-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The Crystal Structure of the Ph Domain of Human Akt3 Protein Kinase
To be Published
|
|
6GTT
 
 | | Human STK10 bound to BIRB-796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Serine/threonine-protein kinase 10 | | Authors: | Berger, B.T, Sorrell, F.J, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | STK10 bound to BIRB-796
To Be Published
|
|
6H5D
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 2 | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
6GLC
 
 | | Structure of phospho-Parkin bound to phospho-ubiquitin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Gladkova, C, Maslen, S.L, Skehel, J.M, Komander, D. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of parkin activation by PINK1.
Nature, 559, 2018
|
|
2XSM
 
 | | Crystal structure of the mammalian cytosolic chaperonin CCT in complex with tubulin | | Descriptor: | CCT | | Authors: | Munoz, I.G, Yebenes, H, Zhou, M, Mesa, P, Serna, M, Bragado-Nilsson, E, Beloso, A, Robinson, C.V, Valpuesta, J.M, Montoya, G. | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structure of the Open Conformation of the Mammalian Chaperonin Cct in Complex with Tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6GR8
 
 | | Human AURKC INCENP complex bound to BRD-7880 | | Descriptor: | 1-[(2~{R},3~{S})-2-[[1,3-benzodioxol-5-ylmethyl(methyl)amino]methyl]-3-methyl-6-oxidanylidene-5-[(2~{S})-1-oxidanylpropan-2-yl]-3,4-dihydro-2~{H}-1,5-benzoxazocin-8-yl]-3-(4-methoxyphenyl)urea, Aurora kinase C, Inner centromere protein | | Authors: | Abdul Azeez, K.R, Sorrell, F.J, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | AURKC INCENP complex bound to BRD-7880
To Be Published
|
|
2XXL
 
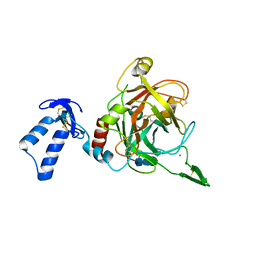 | | Crystal structure of drosophila Grass clip serine protease of Toll pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GRAM-POSITIVE SPECIFIC SERINE PROTEASE, ... | | Authors: | Kellenberger, C, Leone, P, Coquet, L, Jouenne, T, Reichhart, J.M, Roussel, A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analysis of Grass Clip Serine Protease Involved in Drosophila Toll Pathway Activation.
J.Biol.Chem., 286, 2011
|
|
4JXS
 
 | |
2XXS
 
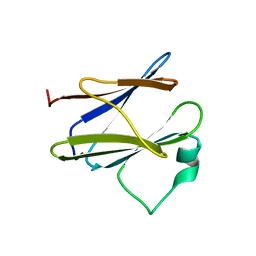 | | Solution structure of the N-terminal domain of the Shigella type III secretion protein MxiG | | Descriptor: | PROTEIN MXIG | | Authors: | McDowell, M.A, Johnson, S, Deane, J.E, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2010-11-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Studies on the N-Terminal Domain of the Shigella Type III Secretion Protein Mxig.
J.Biol.Chem., 286, 2011
|
|
2XVR
 
 | | Phage T7 empty mature head shell | | Descriptor: | MAJOR CAPSID PROTEIN 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Molecular Rearrangements Involved in the Capsid Shell Maturation of Bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
2XGF
 
 | | Structure of the bacteriophage T4 long tail fibre needle-shaped receptor-binding tip | | Descriptor: | CARBONATE ION, FE (II) ION, LONG TAIL FIBER PROTEIN P37 | | Authors: | Bartual, S.G, Otero, J.M, Garcia-Doval, C, Llamas-Saiz, A.L, Kahn, R, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2010-06-03 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacteriophage T4 long tail fiber receptor-binding tip.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2XU6
 
 | | MDV1 coiled coil domain | | Descriptor: | MDV1 COILED COIL | | Authors: | Koirala, S, Bui, H.T, Schubert, H.L, Eckert, D.M, Hill, C.P, Kay, M.S, Shaw, J.M. | | Deposit date: | 2010-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Architecture of a Dynamin Adaptor: Implications for Assembly of Mitochondrial Fission Complexes
J.Cell Biol., 191, 2010
|
|
2XF1
 
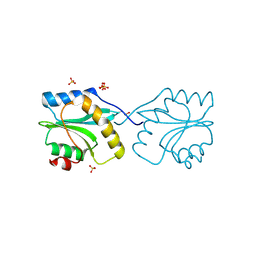 | | Crystal structure of Plasmodium falciparum actin depolymerization factor 1 | | Descriptor: | COFILIN ACTIN-DEPOLYMERIZING FACTOR HOMOLOG 1, SULFATE ION | | Authors: | Singh, B.K, Sattler, J.M, Huttu, J, Chatterjee, M, Schueler, H, Kursula, I. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures Explain Functional Differences in the Two Actin Depolymerization Factors of the Malaria Parasite.
J.Biol.Chem., 286, 2011
|
|
2XDA
 
 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-2-(2-Cyclopropyl)ethyl-4,6,7- trihydroxy-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-2-(2-CYCLOPROPYLETHYL)-4,6,7-TRIHYDROXY-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
