4YGJ
 
 | | NaBr--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | BROMIDE ION, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
4YGN
 
 | | NaI--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | Carbonic anhydrase 2, IODIDE ION, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
6E9V
 
 | | DHF79 filament | | Descriptor: | DHF79 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
5V80
 
 | |
5V84
 
 | | CECR2 in complex with Cpd6 (6-allyl-N,2-dimethyl-7-oxo-N-(1-(1-phenylethyl)piperidin-4-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4-carboxamide) | | Descriptor: | Cat eye syndrome critical region protein 2, N,2-dimethyl-7-oxo-N-{1-[(1S)-1-phenylethyl]piperidin-4-yl}-6-(prop-2-en-1-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4- carboxamide, SULFATE ION | | Authors: | Murray, J.M, Kiefer, J.R, Jayaran, H, Bellon, S, Boy, F. | | Deposit date: | 2017-03-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | GNE-886: A Potent and Selective Inhibitor of the Cat Eye Syndrome Chromosome Region Candidate 2 Bromodomain (CECR2).
ACS Med Chem Lett, 8, 2017
|
|
4YQW
 
 | | Mutant Human DNA Polymerase Eta Q38A/R61A Inserting dCTP Opposite Template G | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Su, Y, Patra, A, Harp, J.M, Egli, M, Guengerich, F.P. | | Deposit date: | 2015-03-13 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | Roles of Residues Arg-61 and Gln-38 of Human DNA Polymerase eta in Bypass of Deoxyguanosine and 7,8-Dihydro-8-oxo-2'-deoxyguanosine.
J.Biol.Chem., 290, 2015
|
|
1XSP
 
 | | Crystal Structure of human DNA polymerase lambda in complex with nicked DNA and pyrophosphate | | Descriptor: | 5'-D(*CP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
4Y6W
 
 | |
1XSN
 
 | | Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap and ddTTP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*(2DT))-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7Z0N
 
 | | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Salgado-Polo, F, Clark, J.M, Macdonald, S.J.F, Barrett, T.N, Perrakis, A, Jamieson, A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators.
J.Med.Chem., 65, 2022
|
|
4YHR
 
 | | Crystal Structure of Yeast Proliferating Cell Nuclear Antigen | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Litman, J.M, Nguyen, V.Q, Kondratick, C.M, Powers, K.T, Schnieders, M.J, Washington, M.T. | | Deposit date: | 2015-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9502 Å) | | Cite: | Dead-End Elimination with a Polarizable Force Field Repacks PCNA Models from Low-Resolution X-ray Diffraction into Atomic Resolution Structures
To be published
|
|
5B4P
 
 | |
7ZAN
 
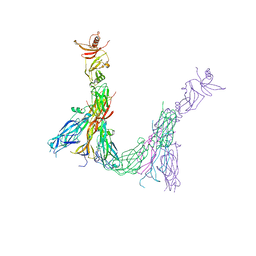 | |
7YX3
 
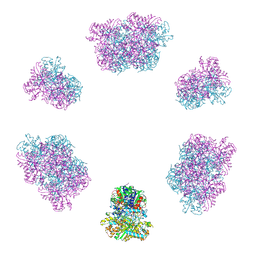 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX4
 
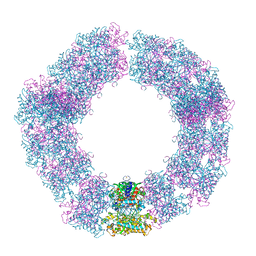 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX5
 
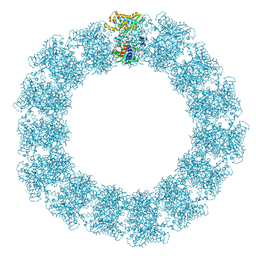 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
2N7A
 
 | | Solution structure of the human Siglec-8 lectin domain | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-07-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1TV9
 
 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE ADENINE AND INCOMING CYTIDINE | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Krahn, J.M, Beard, W.M, Wilson, S.H. | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURAL INSIGHTS INTO DNA POLYMERASE BETA DETERRENTS FOR MISINCORPORATION SUPPORT OF AN INDUCED-FIT MECHANISM FOR FIDELITY
Structure, 12, 2004
|
|
1UCK
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
1TT5
 
 | | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 M, ZINC ION, amyloid protein-binding protein 1, ... | | Authors: | Huang, D.T, Miller, D.W, Mathew, R, Cassell, R, Holton, J.M, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1U85
 
 | |
1UCJ
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
1TOZ
 
 | | NMR structure of the human NOTCH-1 ligand binding region | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hambleton, S, Valeyev, N.Y, Muranyi, A, Knott, V, Werner, J.M, Mcmichael, A.J, Handford, P.A, Downing, A.K. | | Deposit date: | 2004-06-15 | | Release date: | 2004-10-12 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural and functional properties of the human notch-1 ligand binding region
STRUCTURE, 12, 2004
|
|
1U2N
 
 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
1TVA
 
 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE THYMIDINE AND INCOMING CYTIDINE | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*TP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Krahn, J.M, Beard, W.A, Wilson, S.H. | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into DNA polymerase Beta deterrents for misincorporation support an induced-fit mechanism for fidelity
Structure, 12, 2004
|
|
