1BWY
 
 | | NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN | | Descriptor: | PROTEIN (HEART FATTY ACID BINDING PROTEIN) | | Authors: | Lassen, D, Luecke, C, Kveder, M, Mesgarzadeh, A, Schmidt, J.M, Specht, B, Lezius, A, Spener, F, Rueterjans, H. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy.
Eur.J.Biochem., 230, 1995
|
|
1BQI
 
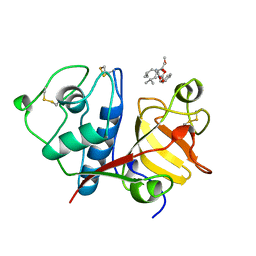 | | USE OF PAPAIN AS A MODEL FOR THE STRUCTURE-BASED DESIGN OF CATHEPSIN K INHIBITORS. CRYSTAL STRUCTURES OF TWO PAPAIN INHIBITOR COMPLEXES DEMONSTRATE BINDING TO S'-SUBSITES. | | Descriptor: | CARBOBENZYLOXY-(L)-LEUCINYL-(L)LEUCINYL METHOXYMETHYLKETONE, PAPAIN | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites.
J.Med.Chem., 41, 1998
|
|
1BQS
 
 | | THE CRYSTAL STRUCTURE OF MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 (MADCAM-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1) | | Authors: | Tan, K, Casasnovas, J.M, Liu, J.H, Briskin, M.J, Springer, T.A, Wang, J.-H. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition.
Structure, 6, 1998
|
|
1BDK
 
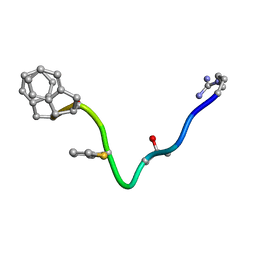 | | AN NMR, CD, MOLECULAR DYNAMICS, AND FLUOROMETRIC STUDY OF THE CONFORMATION OF THE BRADYKININ ANTAGONIST B-9340 IN WATER AND IN AQUEOUS MICELLAR SOLUTIONS | | Descriptor: | bradykinin antagonist B-9340 | | Authors: | Sejbal, J, Kotovych, G, Cann, J.R, Stewart, J.M, Gera, L. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-07 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | An NMR, CD, molecular dynamics, and fluorometric study of the conformation of the bradykinin antagonist B-9340 in water and in aqueous micellar solutions.
J.Med.Chem., 39, 1996
|
|
1C4Z
 
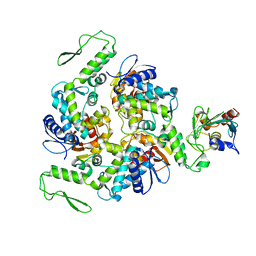 | | STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY | | Descriptor: | UBIQUITIN CONJUGATING ENZYME E2, UBIQUITIN-PROTEIN LIGASE E3A | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
3TDN
 
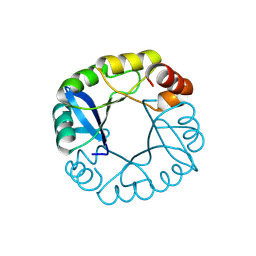 | | Computationally designed two-fold symmetric Tim-barrel protein, FLR | | Descriptor: | FLR SYMMETRIC ALPHA-BETA TIM BARREL | | Authors: | Harp, J.M, Fortenberry, C, Bowman, E, Profitt, W, Dorr, B, Mizoue, L, Meiler, J. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring symmetry as an avenue to the computational design of large protein domains.
J.Am.Chem.Soc., 133, 2011
|
|
1C01
 
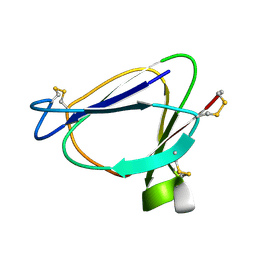 | | SOLUTION STRUCTURE OF MIAMP1, A PLANT ANTIMICROBIAL PROTEIN | | Descriptor: | ANTIMICROBIAL PEPTIDE 1 | | Authors: | McManus, A.M, Nielsen, K.J, Marcus, J.P, Harrison, S.J, Green, J.L, Manners, J.M, Craik, D.J. | | Deposit date: | 1999-07-13 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | MiAMP1, a novel protein from Macadamia integrifolia adopts a Greek key beta-barrel fold unique amongst plant antimicrobial proteins.
J.Mol.Biol., 293, 1999
|
|
1C4B
 
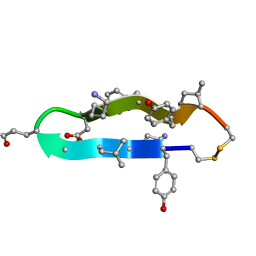 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(RD-262) | | Descriptor: | PROTEIN (CYCLO(RD-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1C3D
 
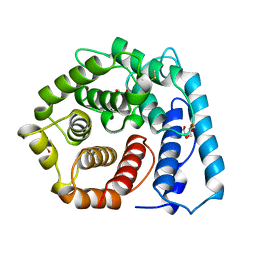 | | X-RAY CRYSTAL STRUCTURE OF C3D: A C3 FRAGMENT AND LIGAND FOR COMPLEMENT RECEPTOR 2 | | Descriptor: | C3D, GLYCEROL | | Authors: | Nagar, B, Jones, R.G, Diefenbach, R.J, Isenman, D.E, Rini, J.M. | | Deposit date: | 1998-05-19 | | Release date: | 1998-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2.
Science, 280, 1998
|
|
1BP4
 
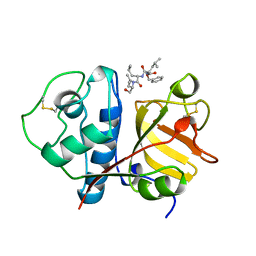 | | USE OF PAPAIN AS A MODEL FOR THE STRUCTURE-BASED DESIGN OF CATHEPSIN K INHIBITORS. CRYSTAL STRUCTURES OF TWO PAPAIN INHIBITOR COMPLEXES DEMONSTRATE BINDING TO S'-SUBSITES. | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide, PAPAIN | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites.
J.Med.Chem., 41, 1998
|
|
1BUE
 
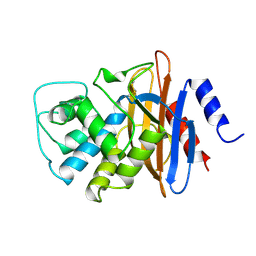 | | NMC-A CARBAPENEMASE FROM ENTEROBACTER CLOACAE | | Descriptor: | PROTEIN (IMIPENEM-HYDROLYSING BETA-LACTAMASE) | | Authors: | Swaren, P, Maveyraud, L, Cabantous, S, Pedelacq, J.D, Mourey, L, Frere, J.M, Samama, J.P. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray analysis of the NMC-A beta-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity.
J.Biol.Chem., 273, 1998
|
|
4LEX
 
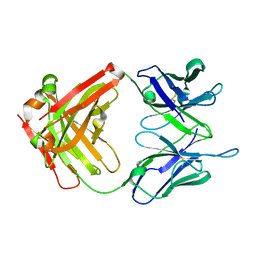 | | Unliganded crystal structure of mAb7 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN | | Authors: | Murray, J.M, Mukund, S. | | Deposit date: | 2013-06-26 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Inhibitory mechanism of an allosteric antibody targeting the glucagon receptor.
J.Biol.Chem., 288, 2013
|
|
4LIM
 
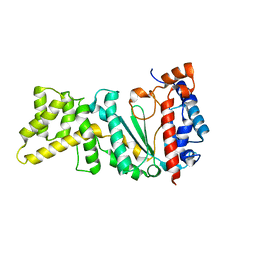 | | Crystal structure of the catalytic subunit of yeast primase | | Descriptor: | DNA primase small subunit, ZINC ION | | Authors: | Vaithiyalingam, S, Chazin, W.J, Berger, J.M, Corn, J, Stephenson, S. | | Deposit date: | 2013-07-02 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into Eukaryotic Primer Synthesis from Structures of the p48 Subunit of Human DNA Primase.
J.Mol.Biol., 426, 2014
|
|
2ZD9
 
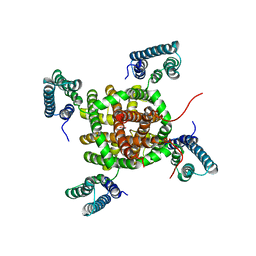 | |
4LCU
 
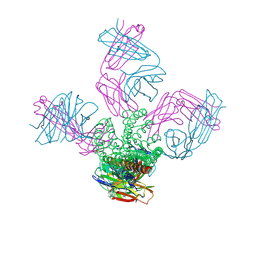 | | Structure of KcsA with E118A mutation | | Descriptor: | DIACYL GLYCEROL, Fab heavy chain, Fab light chain, ... | | Authors: | Nimigean, C.M, Posson, D.J, McCoy, J.M. | | Deposit date: | 2013-06-23 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Molecular interactions involved in proton-dependent gating in KcsA potassium channels.
J.Gen.Physiol., 142, 2013
|
|
4HBT
 
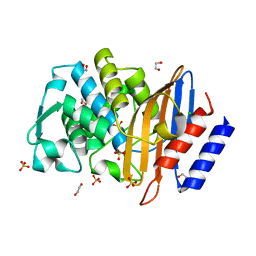 | | Crystal structure of native CTX-M-15 extended-spectrum beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Docquier, J.D, Benvenuti, M, Bruneau, J.M, Rossolini, G.M, Miossec, C, Black, M.T, Mangani, S. | | Deposit date: | 2012-09-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC beta-lactamases
Antimicrob.Agents Chemother., 57, 2013
|
|
1B69
 
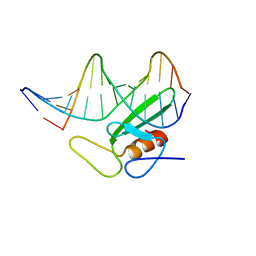 | |
4LF3
 
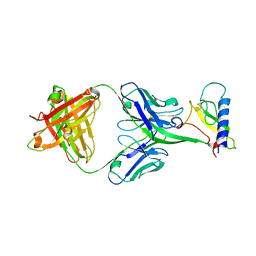 | |
1CBI
 
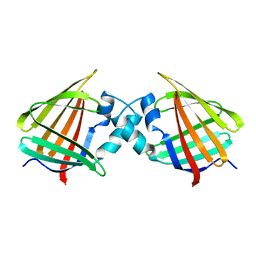 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN I | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN I | | Authors: | Thompson, J.R, Bratt, J.M, Banaszak, L.J. | | Deposit date: | 1995-07-12 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cellular retinoic acid binding protein I shows increased access to the binding cavity due to formation of an intermolecular beta-sheet.
J.Mol.Biol., 252, 1995
|
|
1BRD
 
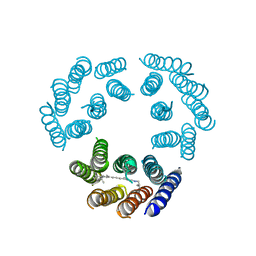 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
1D4V
 
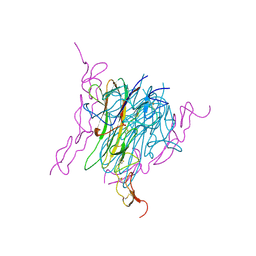 | | Crystal structure of trail-DR5 complex | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND | | Authors: | Mongkolsapaya, J, Grimes, J.M, Stuart, D.I, Jones, E.Y, Screaton, G.R. | | Deposit date: | 1999-10-06 | | Release date: | 1999-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation
Nat.Struct.Biol., 6, 1999
|
|
1D6I
 
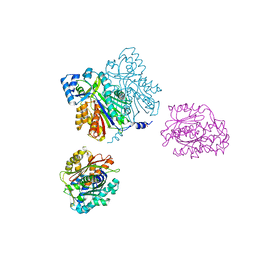 | | CHALCONE SYNTHASE (H303Q MUTANT) | | Descriptor: | CHALCONE SYNTHASE, SULFATE ION | | Authors: | Jez, J.M, Ferrer, J.L, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1999-10-13 | | Release date: | 2000-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
Biochemistry, 39, 2000
|
|
3L16
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3L13
 
 | | Crystal Structures of Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors | | Descriptor: | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, [3-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenyl]methanol | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3L17
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 4-methyl-5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyrimidin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
