1RQY
 
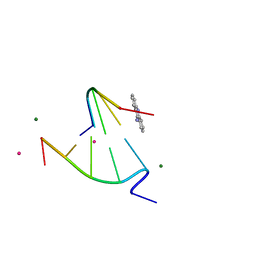 | | 9-amino-[N-(2-dimethylamino)proply]-acridine-4-carboxamide bound to d(CGTACG)2 | | Descriptor: | 5'-D(CP*GP*TP*AP*CP*G)-3', 9-AMINO-N-[3-(DIMETHYLAMINO)PROPYL]ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, ... | | Authors: | Adams, A, Guss, J.M, Denny, W.A, Wakelin, L.P. | | Deposit date: | 2003-12-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 9-amino-[N-(2-dimethylamino)propyl]acridine-4-carboxamide bound to d(CGTACG)(2): a comparison of structures of d(CGTACG)(2) complexed with intercalatorsin the presence of cobalt.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OSV
 
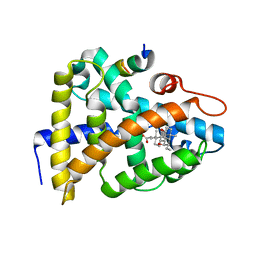 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
2IHN
 
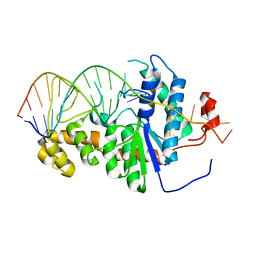 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
4D11
 
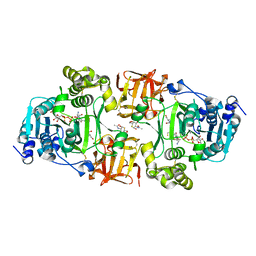 | | GalNAc-T2 crystal soaked with UDP-5SGalNAc, mEA2 peptide and manganese (Lower resolution dataset) | | Descriptor: | 2-acetamido-2-deoxy-5-thio-alpha-D-galactopyranose, MANGANESE (II) ION, PEPTIDE, ... | | Authors: | Lira-Navarrete, E, Iglesias-Fernandez, J, Zandberg, W.F, Companon, I, Kong, Y, Corzana, F, Pinto, B.M, Clausen, H, Peregrina, J.M, Vocadlo, D, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2014-05-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Substrate-Guided Front-Face Reaction Revealed by Combined Structural Snapshots and Metadynamics for the Polypeptide N- Acetylgalactosaminyltransferase 2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3I9F
 
 | | Crystal structure of a putative type 11 methyltransferase from Sulfolobus solfataricus | | Descriptor: | Putative type 11 methyltransferase, ZINC ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative type 11 methyltransferase from Sulfolobus solfataricus
To be Published
|
|
1RRY
 
 | | DHNA complexed with 2-amino-4-hydroxy-5-carboxyethylpyrimidine | | Descriptor: | 2-AMINO-4-HYDROXYPYRIMIDINE-5-CARBOXYLIC ACID ETHYL ESTER, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-09 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
1P1T
 
 | |
2IEN
 
 | | Crystal structure analysis of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
3ENA
 
 | | Crystal structure of the mimivirus NDK +Kpn-N62L-R107G triple mutant complexed with dGDP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-09-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3S9G
 
 | |
2IF6
 
 | | Crystal structure of metalloprotein yiiX from Escherichia coli O157:H7, DUF1105 | | Descriptor: | Hypothetical protein yiiX, UNKNOWN ATOM OR ION | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical metalloprotein yiiX from Escherichia coli O157:H7
To be Published
|
|
1P7B
 
 | | Crystal structure of an inward rectifier potassium channel | | Descriptor: | POTASSIUM ION, integral membrane channel and cytosolic domains | | Authors: | Kuo, A, Gulbis, J.M, Antcliff, J.F, Rahman, T, Lowe, E.D, Zimmer, J, Cuthbertson, J, Ashcroft, F.M, Ezaki, T, Doyle, D.A. | | Deposit date: | 2003-05-01 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structure of the potassium channel KirBac1.1 in the closed state.
Science, 300, 2003
|
|
1TGV
 
 | | Structure of E. coli Uridine Phosphorylase complexed with 5-Fluorouridine and sulfate | | Descriptor: | 5-FLUOROURIDINE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Bu, W, Settembre, E.C, Sanders, J.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of E. coli Uridine Phosphorylase
To be Published, 2004
|
|
1P33
 
 | | Pteridine reductase from Leishmania tarentolae complex with NADPH and MTX | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Zhao, H, Bray, T, Ouellette, M, Zhao, M, Ferre, R.A, Matthews, D, Whiteley, J.M, Varughese, K.I. | | Deposit date: | 2003-04-16 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of pteridine reductase (PTR1) from Leishmania tarentolae.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P44
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | 5-{[4-(9H-FLUOREN-9-YL)PIPERAZIN-1-YL]CARBONYL}-1H-INDOLE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
3I9V
 
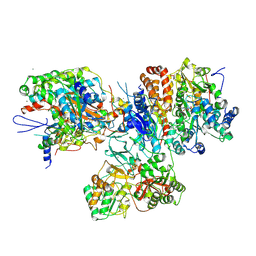 | | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus, oxidized, 2 mol/ASU | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sazanov, L.A, Berrisford, J.M. | | Deposit date: | 2009-07-13 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the mechanism of respiratory complex I
J.Biol.Chem., 284, 2009
|
|
1OMX
 
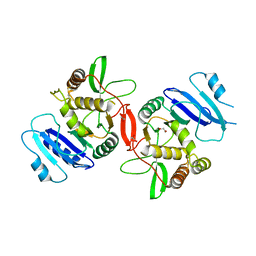 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
4D9U
 
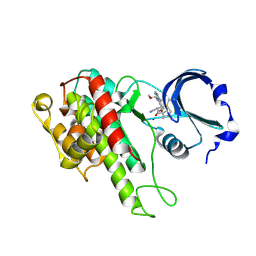 | | Rsk2 C-terminal Kinase Domain, (E)-tert-butyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, tert-butyl (2S)-3-[4-amino-7-(3-hydroxypropyl)-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
1P4Y
 
 | |
3IRV
 
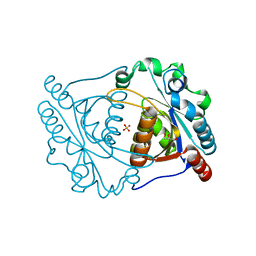 | | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae pv. phaseolicola 1448A | | Descriptor: | CYSTEINE HYDROLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae
To be Published
|
|
3EQZ
 
 | | Crystal structure of a response regulator from Colwellia psychrerythraea | | Descriptor: | Response regulator | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a response regulator from Colwellia psychrerythraea
To be Published
|
|
4CIY
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with (1R,4R,5R)-1,4,5-trihydroxy-3-((1R)-1-hydroxy-2- phenyl)ethylcyclohex-2-en-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[(1R)-1-hydroxy-2-phenyl]ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
1P9P
 
 | | The Crystal Structure of a M1G37 tRNA Methyltransferase, TrmD | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Watts, J.M, Zalacain, M, Van Thiel, A, Vitaszka, P.R, Redlak, M, Andraos-Selim, C, Rastinejad, F, Holmes, W.M. | | Deposit date: | 2003-05-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Catalysis by a Knotted TrmD tRNA Methyltransferase.
J.Mol.Biol., 333, 2003
|
|
2ISQ
 
 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Arabidopsis Thaliana in Complex with C-Terminal Peptide from Arabidopsis Serine Acetyltransferase | | Descriptor: | Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Francois, J.A, Kumaran, S, Jez, J.M. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for interaction of o-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex.
Plant Cell, 18, 2006
|
|
3ISA
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase/isomerase FROM Bordetella parapertussis | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative enoyl-CoA hydratase/isomerase | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF enoyl-CoA hydratase FROM Bordetella parapertussis
To be Published
|
|
