1TOI
 
 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
2IQ1
 
 | | Crystal structure of human PPM1K | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C kappa, PPM1K | | Authors: | Bonanno, J.B, Freeman, J, Russell, M, Bain, K.T, Adams, J, Pelletier, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2IN4
 
 | | Crystal Structure of Myoglobin with Charge Neutralized Heme, ZnDMb-dme | | Descriptor: | Myoglobin, SULFATE ION, ZINC(II)-DEUTEROPORPHYRIN DIMETHYLESTER | | Authors: | Wheeler, K.E, Nocek, J.M, Cull, D.A, Yatsunyk, L.A, Rosenzweig, A.C, Hoffman, B.M. | | Deposit date: | 2006-10-05 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dynamic docking of cytochrome b5 with myoglobin and alpha-hemoglobin: heme-neutralization "squares" and the binding of electron-transfer-reactive configurations.
J.Am.Chem.Soc., 129, 2007
|
|
2IPQ
 
 | | Crystal structure of C-terminal domain of Salmonella Enterica protein STY4665, PFAM DUF1528 | | Descriptor: | Hypothetical protein STY4665 | | Authors: | Ramagopal, U.A, Bonanno, J.B, Gilmore, J, Toro, R, Bain, K.T, Reyes, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of C-terminal domain of Hypothetical protein STY4665
To be Published
|
|
1KU8
 
 | | Crystal structure of Jacalin | | Descriptor: | JACALIN ALPHA CHAIN, JACALIN BETA CHAIN | | Authors: | Bourne, Y, Astoul, C.H, Zamboni, V, Peumans, W.J, Menu-Bouaouiche, L, Van Damme, E.J.M, Barre, A, Rouge, P. | | Deposit date: | 2002-01-21 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose.
Biochem.J., 364, 2002
|
|
7BDU
 
 | |
7BEE
 
 | |
7B8W
 
 | | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropanoylamino)-~{N}-[2-[(phenylmethyl)-[4-(phenylsulfamoyl)phenyl]carbonyl-amino]ethyl]-1,3-thiazole-5-carboxamide, LIM domain kinase 1 | | Authors: | Lee, H, Yosaatmadja, Y, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Elkins, J.M. | | Deposit date: | 2020-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470
To Be Published
|
|
6RMT
 
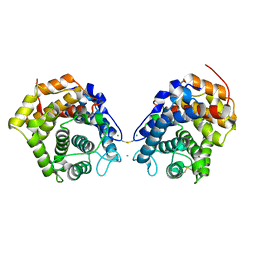 | |
2LKX
 
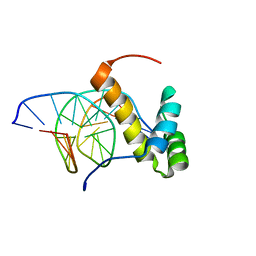 | | NMR structure of the homeodomain of Pitx2 in complex with a TAATCC DNA binding site | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3'), Pituitary homeobox 3 | | Authors: | Baird-Titus, J.M, Doerdelmann, T, Chaney, B.A, Clark-Baldwin, K, Dave, V, Ma, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome
Biochemistry, 44, 2005
|
|
7CW3
 
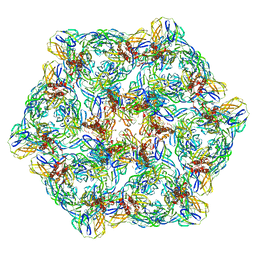 | | Cryo-EM structure of Chikungunya virus in complex with mAb CHK-263 IgG (subregion around icosahedral 2-fold vertex) | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1AJ8
 
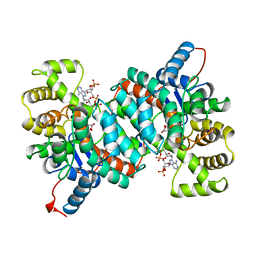 | | CITRATE SYNTHASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | CITRATE SYNTHASE, CITRIC ACID, COENZYME A | | Authors: | Russell, R.J.M, Ferguson, J.M.C, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolution,.
Biochemistry, 36, 1997
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7CN9
 
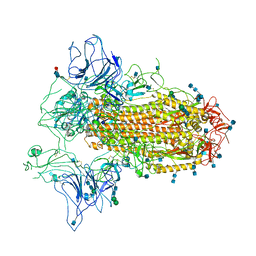 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
7CW0
 
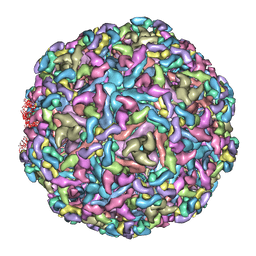 | | Cryo-EM structure of Chikungunya virus in complex with mAb CHK-263 IgG | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RMU
 
 | | Crystal structure of disulphide-linked human C3d dimer in complex with Staphylococcus aureus complement subversion protein Sbi-IV | | Descriptor: | 1,2-ETHANEDIOL, Complement C3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wahid, A.A, van den Elsen, J.M.H, Crennell, S.J. | | Deposit date: | 2019-05-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Staphylococcal Complement Evasion Protein Sbi Stabilises C3d Dimers by Inducing an N-Terminal Helix Swap
Front Immunol, 13, 2022
|
|
1MPA
 
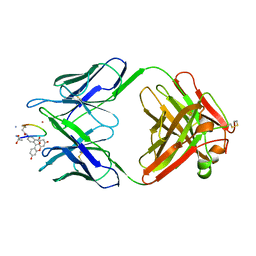 | | BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS | | Descriptor: | CADMIUM ION, MN12H2 IGG2A-KAPPA, PORA P1.16 PEPTIDE FLUORESCEIN CONJUGATE | | Authors: | Van Den Elsen, J.M.H, Herron, J.N, Kroon, J, Gros, P. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bactericidal antibody recognition of a PorA epitope of Neisseria meningitidis: crystal structure of a Fab fragment in complex with a fluorescein-conjugated peptide.
Proteins, 29, 1997
|
|
7CW2
 
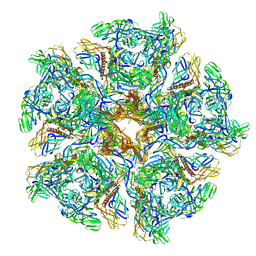 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 (subregion around icosahedral 5-fold vertex) | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CVZ
 
 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CVY
 
 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-124 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1MNT
 
 | | SOLUTION STRUCTURE OF DIMERIC MNT REPRESSOR (1-76) | | Descriptor: | MNT REPRESSOR | | Authors: | Burgering, M.J.M, Boelens, R, Gilbert, D.E, Breg, J.N, Knight, K.L, Sauer, R.T, Kaptein, R. | | Deposit date: | 1994-06-28 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dimeric Mnt repressor (1-76).
Biochemistry, 33, 1994
|
|
6SAL
 
 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM26 | | Descriptor: | 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Doveston, R.G, Brunsveld, L. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Ligand-Based Design of Allosteric Retinoic Acid Receptor-Related Orphan Receptor gamma t (ROR gamma t) Inverse Agonists.
J.Med.Chem., 63, 2020
|
|
7CUE
 
 | | Crystal structure of HID2 bound to human Hemoglobin | | Descriptor: | Amino acid ABC transporter substrate-binding protein, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Caaveiro, J.M.M, Hoshino, M, Tsumoto, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the recognition of human hemoglobin by the Shr protein from Streptococcus pyogenes
To Be Published
|
|
7CUD
 
 | |
7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
