1Q52
 
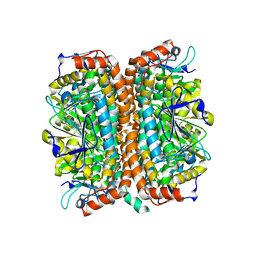 | | Crystal Structure of Mycobacterium tuberculosis MenB, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
1VYD
 
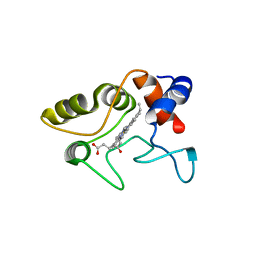 | | Crystal structure of cytochrome C2 mutant G95E | | Descriptor: | CYTOCHROME C2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dumortier, C, Fitch, J, Van Petegem, F, Vermeulen, W, Meyer, T.E, Van Beeumen, J.J, Cusanovich, M.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-06-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein Dynamics in the Region of the Sixth Ligand Methionine Revealed by Studies of Imidazole Binding to Rhodobacter Capsulatus Cytochrome C2 Hinge Mutants.
Biochemistry, 43, 2004
|
|
2YP9
 
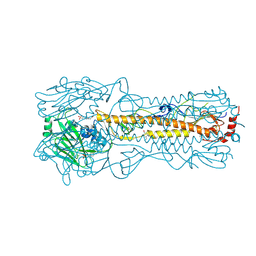 | | Haemagglutinin of 2005 Human H3N2 Virus in Complex with Avian Receptor Analogue 3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP7
 
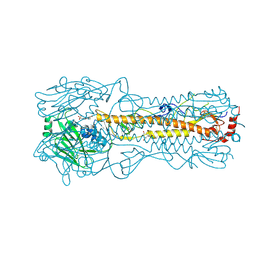 | | Haemagglutinin of 2005 Human H3N2 Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1QSC
 
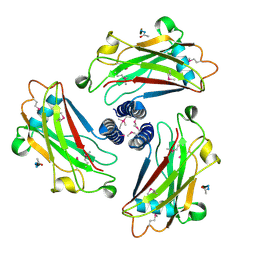 | | CRYSTAL STRUCTURE OF THE TRAF DOMAIN OF TRAF2 IN A COMPLEX WITH A PEPTIDE FROM THE CD40 RECEPTOR | | Descriptor: | CD40 RECEPTOR, TNF RECEPTOR ASSOCIATED FACTOR 2 | | Authors: | McWhirter, S.M, Pullen, S.S, Holton, J.M, Crute, J.J, Kehry, M.R, Alber, T. | | Deposit date: | 1999-06-20 | | Release date: | 1999-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis of CD40 recognition and signaling by human TRAF2.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3FST
 
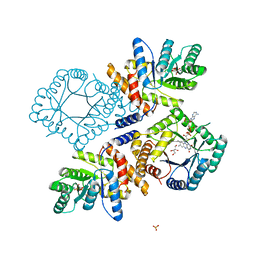 | |
1N2R
 
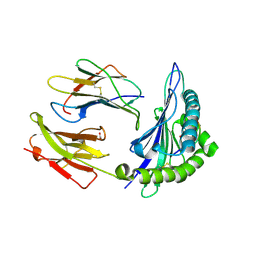 | | A natural selected dimorphism in HLA B*44 alters self, peptide reportoire and T cell recognition. | | Descriptor: | ACETIC ACID, Beta-2-microglobulin, HLA DPA*0201 PEPTIDE, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-10-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
3FSU
 
 | | Crystal Structure of Escherichia coli Methylenetetrahydrofolate Reductase Double Mutant Phe223LeuGlu28Gln complexed with methyltetrahydrofolate | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional role for the conformationally mobile phenylalanine 223 in the reaction of methylenetetrahydrofolate reductase from Escherichia coli.
Biochemistry, 48, 2009
|
|
1X86
 
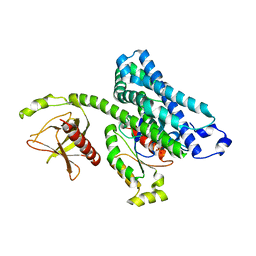 | |
1M6O
 
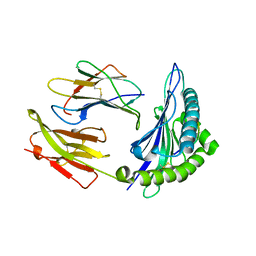 | | Crystal Structure of HLA B*4402 in complex with HLA DPA*0201 peptide | | Descriptor: | Beta-2-microglobulin, HLA DPA*0201 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N.A, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-07-17 | | Release date: | 2003-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1MIT
 
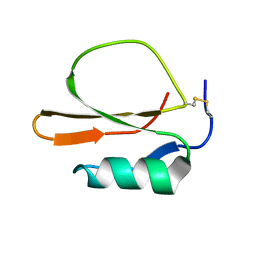 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1ZUQ
 
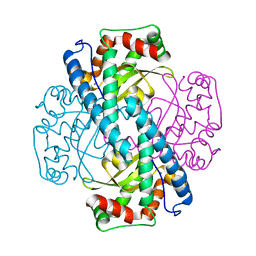 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Hearn, A.S, Perry, J.J, Cabelii, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-31 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
1ZSP
 
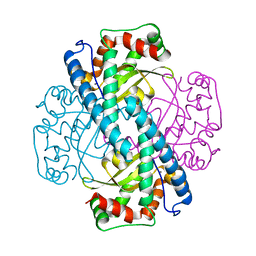 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Hearn, A.S, Perry, J.J, Cabelli, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
1MDF
 
 | | CRYSTAL STRUCTURE OF DhbE IN ABSENCE OF SUBSTRATE | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase, SULFATE ION | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1ZSD
 
 | | Crystal Structure Of HLA-B*3501 Presenting an 11-Mer EBV Antigen EPLPQGQLTAY | | Descriptor: | BZLF1 trans-activator protein, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Miles, J.J, Elhassen, D, Borg, N.A, Silins, S.L, Tynan, F.E, Burrows, J.M, Purcell, A.W, Kjer-Nielsen, L, Rossjohn, J, Burrows, S.R, McCluskey, J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CTL Recognition of a Bulged Viral Peptide Involves Biased TCR Selection.
J.Immunol., 175, 2005
|
|
3B96
 
 | | Structural Basis for Substrate Fatty-Acyl Chain Specificity: Crystal Structure of Human Very-Long-Chain Acyl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TETRADECANOYL-COA, Very long-chain specific acyl-CoA dehydrogenase | | Authors: | McAndrew, R.P, Wang, Y, Mohsen, A.W, He, M, Vockley, J, Kim, J.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3BB7
 
 | | Structure of Prevotella intermedia prointerpain A fragment 39-359 (mutant C154A) | | Descriptor: | interpain A | | Authors: | Mallorqui-Fernandez, N, Manandhar, S.P, Mallorqui-Fernandez, G, Uson, I, Wawrzonek, K, Kantyka, T, Sola, M, Thogersen, I.B, Enghild, J.J, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A New Autocatalytic Activation Mechanism for Cysteine Proteases Revealed by Prevotella intermedia Interpain A
J.Biol.Chem., 283, 2008
|
|
1ZTE
 
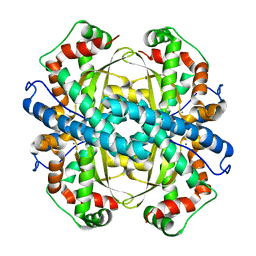 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Suerpoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Hearn, A.S, Perry, J.J, Cabelii, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-26 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
1ZZA
 
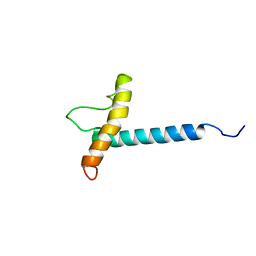 | | Solution NMR Structure of the Membrane Protein Stannin | | Descriptor: | Stannin | | Authors: | Buck-Koehntop, B.A, Mascioni, A, Buffy, J.J, Veglia, G. | | Deposit date: | 2005-06-13 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and membrane topology of stannin: a mediator of neuronal cell apoptosis induced by trimethyltin chloride.
J.Mol.Biol., 354, 2005
|
|
3BBA
 
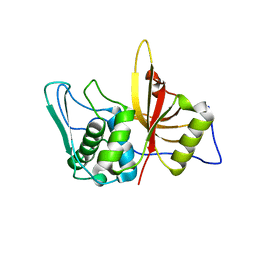 | | Structure of active wild-type Prevotella intermedia interpain A cysteine protease | | Descriptor: | interpain A | | Authors: | Mallorqui-Fernandez, N, Manandhar, S.P, Mallorqui-Fernandez, G, Uson, I, Wawrzonek, K, Kantyka, T, Sola, M, Thogersen, I.B, Enghild, J.J, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A New Autocatalytic Activation Mechanism for Cysteine Proteases Revealed by Prevotella intermedia Interpain A
J.Biol.Chem., 283, 2008
|
|
1ZXA
 
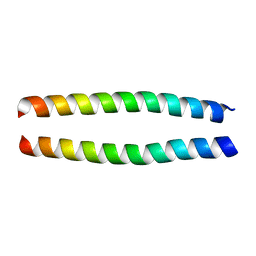 | | Solution Structure of the Coiled-Coil Domain of cGMP-dependent Protein Kinase Ia | | Descriptor: | cGMP-dependent protein kinase 1, alpha isozyme | | Authors: | Schnell, J.R, Zhou, G.P, Zweckstetter, M, Rigby, A.C, Chou, J.J. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rapid and accurate structure determination of coiled-coil domains using NMR dipolar couplings: Application to cGMP-dependent protein kinase I{alpha}
Protein Sci., 14, 2005
|
|
1ZYR
 
 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
2AAG
 
 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | Malonate Semialdehyde Decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
3L51
 
 | | Crystal Structure of the Mouse Condensin Hinge Domain | | Descriptor: | GLYCEROL, Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Griese, J.J, Hopfner, K.-P. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins
Nucleic Acids Res., 38, 2010
|
|
3TG0
 
 | | E. coli alkaline phosphatase with bound inorganic phosphate | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bobyr, E, Lassila, J.K, Wiersma-Koch, H.I, Fenn, T.D, Lee, J.J, Nikolic-Hughes, I, Hodgson, K.O, Rees, D.C, Hedman, B, Herschlag, D. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution analysis of Zn(2+) coordination in the alkaline phosphatase superfamily by EXAFS and x-ray crystallography.
J.Mol.Biol., 415, 2012
|
|
