4AVZ
 
 | | Tailspike protein mutant E372Q of E. coli bacteriophage HK620 | | Descriptor: | TAIL SPIKE PROTEIN, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Gohlke, U, Broeker, N.K, Mueller, J.J, Uetrecht, C, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
7L0P
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4WZX
 
 | | ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins | | Descriptor: | COBALT (II) ION, IST1 homolog, SULFATE ION, ... | | Authors: | Caballe, A, Wenzel, D.M, Agromayor, M, Alam, S.L, Skalicky, J.J, Kloc, M, Carlton, J.G, Labrador, L, Sundquist, W.I, Martin-Serrano, J. | | Deposit date: | 2014-11-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3859 Å) | | Cite: | ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins.
Elife, 4, 2015
|
|
1XF3
 
 | | Structure of ligand-free Fab DNA-1 in space group P65 | | Descriptor: | Fab Light chain, Fab heavy chain | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
7L0S
 
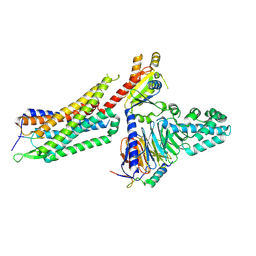 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3E7X
 
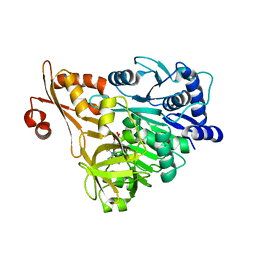 | | Crystal structure of DLTA: implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1 | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
7NA0
 
 | |
1ONB
 
 | |
7MYB
 
 | |
1XDC
 
 | | Hydrogen Bonding in Human Manganese Superoxide Dismutase containing 3-Fluorotyrosine | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Ayala, I, Perry, J.J, Szczepanski, J, Cabelli, D.E, Tainer, J.A, Vala, M.T, Nick, H.S, Silverman, D.N. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydrogen bonding in human manganese superoxide dismutase containing 3-fluorotyrosine
Biophys.J., 89, 2005
|
|
7MY9
 
 | | Structure of proline utilization A with 1,3-dithiolane-2-carboxylate bound in the proline dehydrogenase active site | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Campbell, A.C. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Photoinduced Covalent Irreversible Inactivation of Proline Dehydrogenase by S-Heterocycles.
Acs Chem.Biol., 16, 2021
|
|
7MYA
 
 | |
7MYC
 
 | |
1XF4
 
 | | Structure of ligand-free Fab DNA-1 in space group P321 solved from crystals with perfect hemihedral twinning | | Descriptor: | Fab heavy chain, Fab light chain, SULFATE ION | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
1NQD
 
 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM HISTOLYTICUM COLG COLLAGENASE COLLAGEN-BINDING DOMAIN 3B AT 1.65 ANGSTROM RESOLUTION IN PRESENCE OF CALCIUM | | Descriptor: | CALCIUM ION, class 1 collagenase | | Authors: | Wilson, J.J, Matsushita, O, Okabe, A, Sakon, J. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Bacterial Collagen-Binding Domain with Novel Calcium-Binding Motif Controls Domain Orientation
Embo J., 22, 2003
|
|
3BWT
 
 | |
1XLS
 
 | | Crystal structure of the mouse CAR/RXR LBD heterodimer bound to TCPOBOP and 9cRA and a TIF2 peptide containg the third LXXLL motifs | | Descriptor: | (9cis)-retinoic acid, 3,5-DICHLORO-2-{4-[(3,5-DICHLOROPYRIDIN-2-YL)OXY]PHENOXY}PYRIDINE, Nuclear receptor coactivator 2, ... | | Authors: | Suino, K, peng, L, Reynolds, R, Li, Y, Cha, J.-Y, Repa, J.J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2004-09-30 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization.
Mol.Cell, 16, 2004
|
|
1W7T
 
 | |
7MWT
 
 | |
7MWV
 
 | |
7MWU
 
 | |
1BDM
 
 | |
3CP1
 
 | |
3CPM
 
 | | plant peptide deformylase PDF1B crystal structure | | Descriptor: | Peptide deformylase, chloroplast, SULFATE ION, ... | | Authors: | Rodgers, D.W, Houtz, R.L, Dirk, L.M.A, Schmidt, J.J, Cai, Y. | | Deposit date: | 2008-03-31 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the substrate specificity of plant peptide deformylase, an essential enzyme with potential for the development of novel biotechnology applications in agriculture
Biochem.J., 413, 2008
|
|
4ZVW
 
 | | Structure of apo human ALDH7A1 in space group C2 | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J. | | Deposit date: | 2015-05-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
