8UAP
 
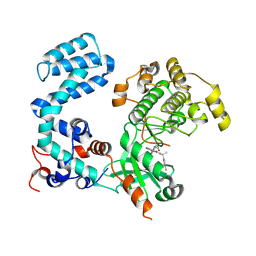 | | Crystal Structure of Human G Protein-Coupled Receptor Kinase 5 D311N in Complex with CCG273441 | | Descriptor: | (3Z)-3-{[4-(2-chloroacetamido)-3,5-dimethyl-1H-pyrrol-2-yl]methylidene}-N-[(1R)-1-(4-fluorophenyl)ethyl]-2-oxo-2,3-dihydro-1H-indole-5-carboxamide, G protein-coupled receptor kinase 5 | | Authors: | Chen, Y, Tesmer, J.J.G. | | Deposit date: | 2023-09-21 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a new class of potent and highly selective G protein-coupled receptor kinase 5 inhibitors and structural insight from crystal structures of inhibitor complexes.
Eur.J.Med.Chem., 264, 2023
|
|
8UAQ
 
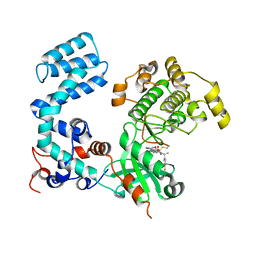 | | Crystal Structure of Human G Protein-Coupled Receptor Kinase 5 in Complex with GRL018-21 | | Descriptor: | (3Z)-N-[(1R)-1-(4-fluorophenyl)ethyl]-3-[(4-{[(2S)-2-(furan-2-yl)-2-hydroxyacetyl]amino}-3,5-dimethyl-1H-pyrrol-2-yl)methylidene]-2-oxo-2,3-dihydro-1H-indole-5-carboxamide, G protein-coupled receptor kinase 5 | | Authors: | Chen, Y, Tesmer, J.J.G. | | Deposit date: | 2023-09-21 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a new class of potent and highly selective G protein-coupled receptor kinase 5 inhibitors and structural insight from crystal structures of inhibitor complexes.
Eur.J.Med.Chem., 264, 2023
|
|
4ECE
 
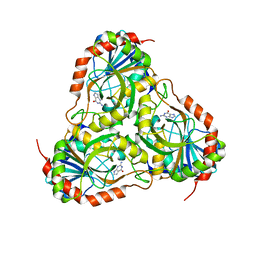 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with guanine | | Descriptor: | GUANINE, Purine nucleoside phosphorylase | | Authors: | Haapalainen, A.M, Ho, M.C, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-26 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
2GDS
 
 | |
1ZO8
 
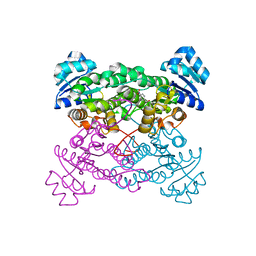 | | X-ray Structure of the haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (S)-para-nitrostyrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (S)-PARA-NITROSTYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Tang, L, Villa, A, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
2GDF
 
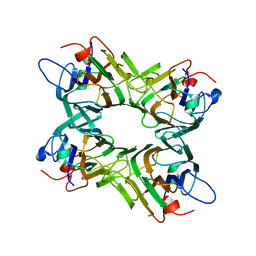 | |
1EFV
 
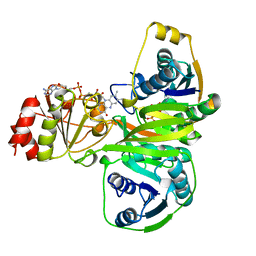 | |
1YY9
 
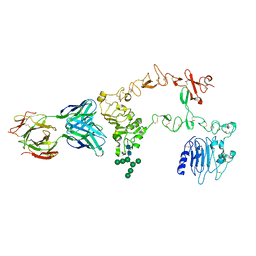 | | Structure of the extracellular domain of the epidermal growth factor receptor in complex with the Fab fragment of cetuximab/Erbitux/IMC-C225 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab Heavy chain, ... | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JYB
 
 | | Binary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JW9
 
 | | Ternary cocrystal structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, SODIUM ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JV3
 
 | |
7K64
 
 | | Binary titrated soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
4EB8
 
 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Ho, M.C, Haapalainen, A.M, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
1YY8
 
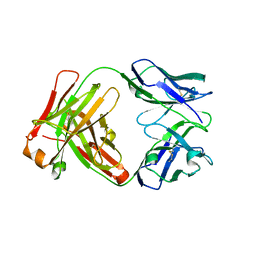 | | Crystal structure of the Fab fragment from the monoclonal antibody cetuximab/Erbitux/IMC-C225 | | Descriptor: | Cetuximab Fab Heavy chain, Cetuximab Fab Light chain | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
1CJK
 
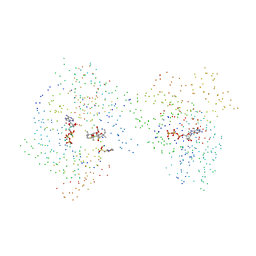 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH ADENOSINE 5'-(ALPHA THIO)-TRIPHOSPHATE (RP), MG, AND MN | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADENOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1ZAE
 
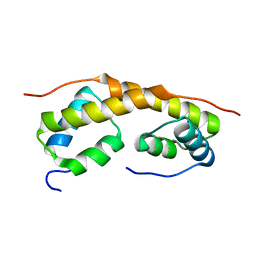 | | Solution structure of the functional domain of phi29 replication organizer p16.7c | | Descriptor: | Early protein GP16.7 | | Authors: | Asensio, J.L, Albert, A, Munoz-Espin, D, Gonzalez, C, Hermoso, J, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional domain of phi29 replication organizer: insights into oligomerization and dna binding
J.Biol.Chem., 280, 2005
|
|
4EAR
 
 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Haapalainen, A.M, Ho, M.C, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-22 | | Release date: | 2013-02-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
1ZCA
 
 | |
1DHG
 
 | | HG-SUBSTITUTED DESULFOREDOXIN | | Descriptor: | MERCURY (II) ION, PROTEIN (DESULFOREDOXIN) | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Moura, I, Moura, J.J.G, Rusnak, F, Romao, M.J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1E08
 
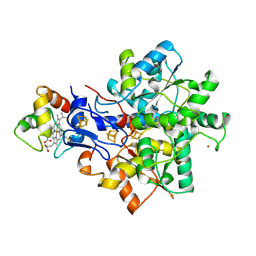 | | Structural model of the [Fe]-Hydrogenase/cytochrome c553 complex combining NMR and soft-docking | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Morelli, X, Czjzek, M, Hatchikian, C.E, Bornet, O, Fontecilla-Camps, J.C, Palma, N.P, Moura, J.J.G, Guerlesquin, F. | | Deposit date: | 2000-03-13 | | Release date: | 2000-08-25 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
J.Biol.Chem., 275, 2000
|
|
7MTB
 
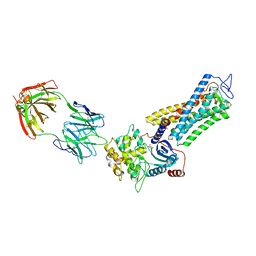 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab6 | | Descriptor: | Fab6 heavy chain, Fab6 light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT8
 
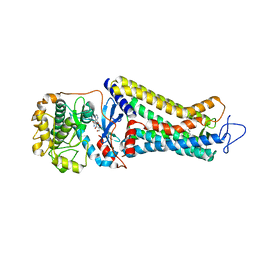 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MTA
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab1 | | Descriptor: | Fab1 Heavy chain, Fab1 Light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT9
 
 | | Rhodopsin kinase (GRK1) in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
