6JFL
 
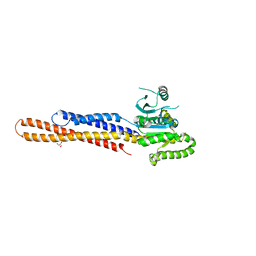 | | Nucleotide-free Mitofusin2 (MFN2) | | Descriptor: | CALCIUM ION, GLYCEROL, Mitofusin-2,cDNA FLJ57997, ... | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
2I33
 
 | |
6JFM
 
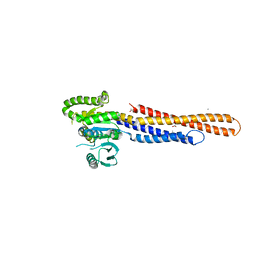 | | Mitofusin2 (MFN2)_T111D | | Descriptor: | ACETATE ION, CALCIUM ION, Mitofusin-2,Mitofusin-2 | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
5KEK
 
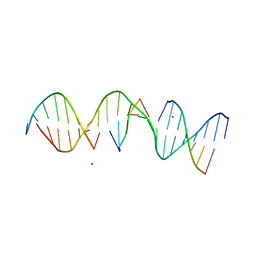 | | Structure Determination of a Self-Assembling DNA Crystal | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, Birktoft, J.J, Seeman, N.C, Yan, H. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Construction and Structure Determination of a Three-Dimensional DNA Crystal.
J.Am.Chem.Soc., 138, 2016
|
|
5KF6
 
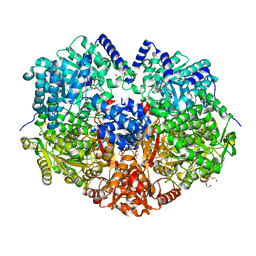 | |
6JMA
 
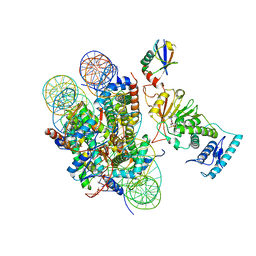 | | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | | Descriptor: | DNA I&J, Histone H2A, Histone H2B 1.1, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
6JQ0
 
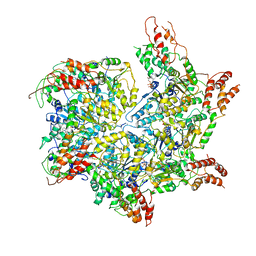 | | CryoEM structure of Abo1 Walker B (E372Q) mutant hexamer - ATP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Uncharacterized AAA domain-containing protein C31G5.19, ... | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
5KEO
 
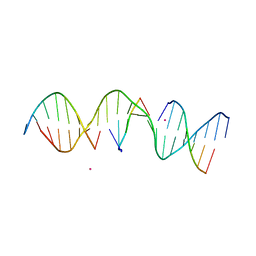 | | Structure Determination of a Self-Assembling DNA Crystal | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, Birktoft, J.J, Seeman, N.C, Yan, H. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Construction and Structure Determination of a Three-Dimensional DNA Crystal.
J.Am.Chem.Soc., 138, 2016
|
|
6JCE
 
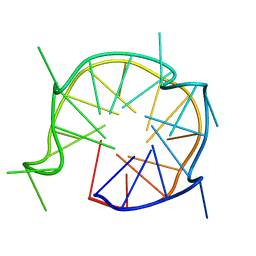 | | NMR solution and X-ray crystal structures of a DNA containing both right-and left-handed parallel-stranded G-quadruplexes | | Descriptor: | 29-mer DNA | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
2LZY
 
 | |
2FBV
 
 | | WRN exonuclease, Mn complex | | Descriptor: | MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5K8S
 
 | | cAMP bound PfPKA-R (297-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
2MDZ
 
 | | NMR structure of the Paracoccus denitrificans Z-subunit determined in the presence of ADP | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Wuthrich, K, Morales-Rios, E, Zarco-Zavala, M, Garcia-Trejo, J.J, Dutta, S.K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans
To be Published
|
|
2FBT
 
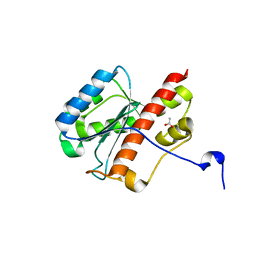 | | WRN exonuclease | | Descriptor: | ACETIC ACID, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2LVI
 
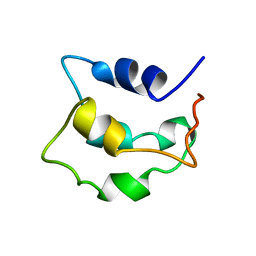 | | Solution structure of apo-Phl p 7 | | Descriptor: | Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
8OQ2
 
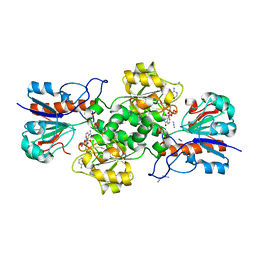 | | Binding of NADP to a formate dehydrogenase from Starkeya novella. | | Descriptor: | AZIDE ION, Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Partipilo, M, Whittaker, J.J, Pontillo, N, Guskov, A, Slotboom, D.J. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Binding of NADP to a formate dehydrogenase from Starkeya novella.
To Be Published
|
|
2LXM
 
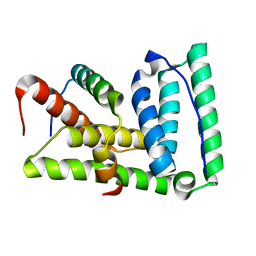 | | Lip5-chmp5 | | Descriptor: | Charged multivesicular body protein 5, Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions of the Human LIP5 Regulatory Protein with Endosomal Sorting Complexes Required for Transport.
J.Biol.Chem., 287, 2012
|
|
5KBZ
 
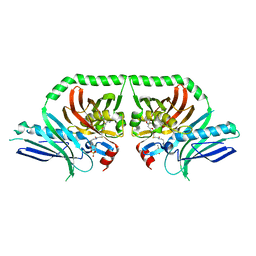 | | Structure of the PksA Product Template domain in complex with a phosphopantetheine mimetic | | Descriptor: | (14R)-14-hydroxy-15,15-dimethyl-1-[5-({[(5-methyl-1,2-oxazol-3-yl)methyl]sulfanyl}methyl)-1,2-oxazol-3-yl]-4,9,13-trioxo-2-thia-5,8,12-triazahexadecan-16-yl dihydrogen phosphate, Noranthrone synthase | | Authors: | Tsai, S.C, Burkart, M.D, Townsend, C.A, Barajas, J.F, Shakya, G, Topper, C.L, Moreno, G, Jackson, D.R, Rivera, H, La Clair, J.J, Vagstad, A. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Polyketide mimetics yield structural and mechanistic insights into product template domain function in nonreducing polyketide synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2MAH
 
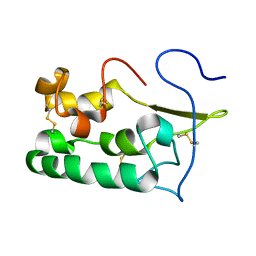 | | Solution structure of Smoothened | | Descriptor: | Protein smoothened | | Authors: | Rana, R, Lee, H, Zheng, J.J. | | Deposit date: | 2013-07-09 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling.
Nat Commun, 4, 2013
|
|
2LZX
 
 | |
2GVA
 
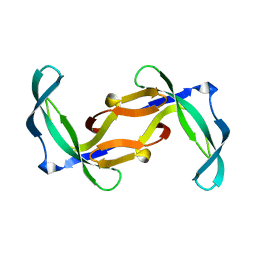 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
2GVB
 
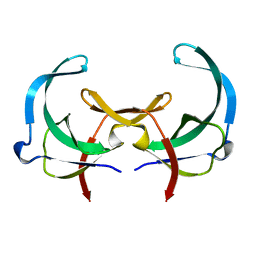 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
8P3C
 
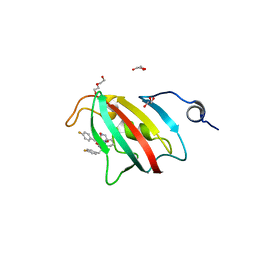 | | Full length structure of BpMIP with bound inhibitor NJS227. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-(pyridin-3-ylmethylamino)propan-2-yl]piperidine-2-carboxamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
8P3D
 
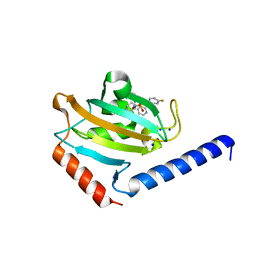 | | Full length structure of TcMIP with bound inhibitor NJS224. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-4-methyl-1-oxidanylidene-1-(pyridin-3-ylmethylamino)pentan-2-yl]piperidine-2-carboxamide, SODIUM ION, peptidylprolyl isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
8P42
 
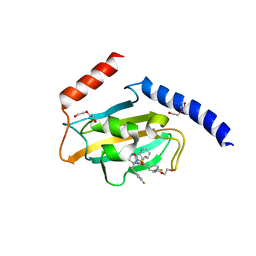 | | Full length structure of TcMIP with bound inhibitor NJS227. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-(pyridin-3-ylmethylamino)propan-2-yl]piperidine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Macrophage infectivity potentiator | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-19 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
