6T8I
 
 | | Crystal structure of wild type EndoBT-3987 from Bacteroides thetaiotamicron VPI-5482 | | Descriptor: | Endo-beta-N-acetylglucosaminidase F1, GLYCEROL | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6T8L
 
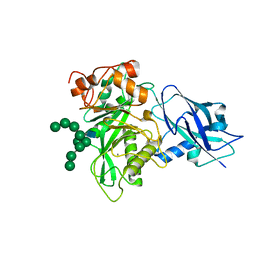 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man9GlcNAc product in P212121 | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
2YIL
 
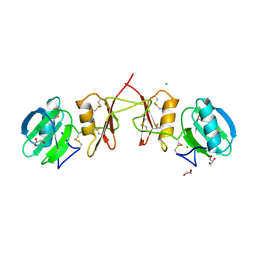 | | Crystal Structure of Parasite Sarcocystis muris Lectin SML-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, MICRONEME ANTIGEN L2, ... | | Authors: | Mueller, J.J, Weiss, M.S, Heinemann, U. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pan-Modular Structure of Microneme Protein Sml-2 from Parasite Sarcocystis Muris at 1.95 A Resolution and its Complex with 1-Thio-Beta-D-Galactose.
Acta Crystallogr.,Sect.D, D67, 2011
|
|
4XSJ
 
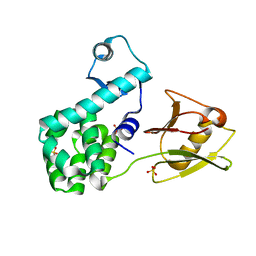 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
6TJY
 
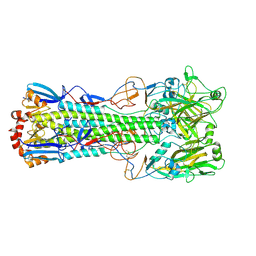 | | Crystal structure of haemagglutinin from (A/seal/Germany/1/2014) seal H10N7 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6TCW
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man5GlcNAc product | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
3AVH
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
6TJW
 
 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Del228) from an H10N7 seal influenza virus isolated in Germany | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
3AVL
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
4XTB
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
2CW0
 
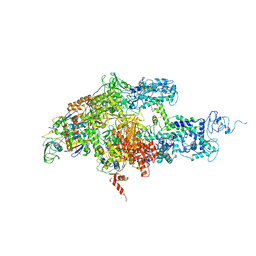 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
3AVN
 
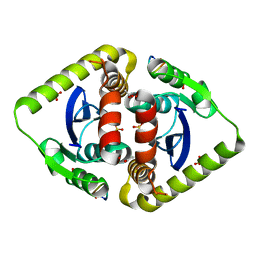 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVA
 
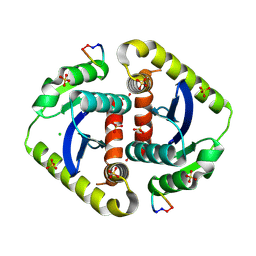 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVI
 
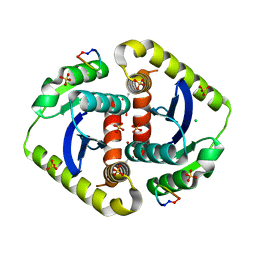 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
5W93
 
 | | p130Cas complex with paxillin LD1 | | Descriptor: | Breast cancer anti-estrogen resistance protein 1, Paxillin | | Authors: | Miller, D.J, Zheng, J.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and functional insights into the interaction between the Cas family scaffolding protein p130Cas and the focal adhesion-associated protein paxillin.
J. Biol. Chem., 292, 2017
|
|
3AVB
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVJ
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AV9
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
5WFE
 
 | | Cas1-Cas2-IHF-DNA holo-complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Wright, A.V, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
7US3
 
 | | Structure of Putrescine N-hydroxylase Involved Complexed with NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putrescine N-hydroxylase, ... | | Authors: | Tanner, J.J, Bogner, A.N. | | Deposit date: | 2022-04-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N -Hydroxylase from Acinetobacter baumannii.
Biochemistry, 61, 2022
|
|
3AVF
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
6TVC
 
 | | Crystal structure of the haemagglutinin from a transmissible H10N7 seal influenza virus isolated in Netherland | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Haemagglutinin HA1, Haemagglutinin HA2 | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6TVT
 
 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Del228) from an H10N7 seal influenza virus isolated in Germany in complex with human receptor analogue 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-10 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6TXO
 
 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Del228) from an H10N7 seal influenza virus isolated in Germany in complex with avian receptor analogue 3'-SLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
2BBU
 
 | |
