2C1J
 
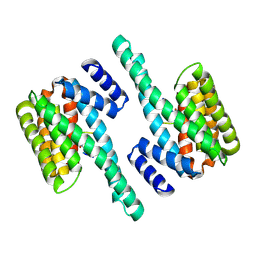 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | 分子名称: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | 著者 | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | 登録日 | 2005-09-15 | | 公開日 | 2005-11-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
2C0S
 
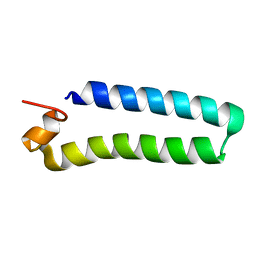 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | 分子名称: | CONSERVED DOMAIN PROTEIN | | 著者 | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | 登録日 | 2005-09-07 | | 公開日 | 2006-09-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2C40
 
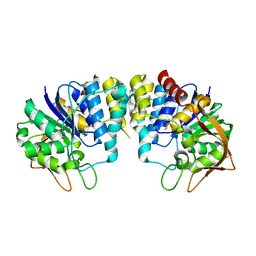 | | CRYSTAL STRUCTURE OF INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FROM BACILLUS ANTHRACIS AT 2.2A RESOLUTION | | 分子名称: | CALCIUM ION, INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FAMILY PROTEIN, alpha-D-ribofuranose | | 著者 | Moroz, O.V, Blagova, E.V, Fogg, M.J, Levdikov, V.M, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | 登録日 | 2005-10-13 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Inosine-Uridine Preferring Nucleoside Hydrolase from Bacillus Anthracis at 2.2A Resolution
To be Published
|
|
2BJH
 
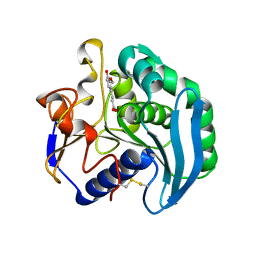 | | Crystal Structure Of S133A AnFaeA-ferulic acid complex | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | 著者 | Faulds, C.B, Molina, R, Gonzalez, R, Husband, F, Juge, N, Sanz-Aparicio, J, Hermoso, J.A. | | 登録日 | 2005-02-02 | | 公開日 | 2005-09-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Probing the Determinants of Substrate Specificity of a Feruloyl Esterase, Anfaea, from Aspergillus Niger
FEBS J., 272, 2005
|
|
3Q7P
 
 | |
7KMU
 
 | | Structure of WT Malaysian Banana Lectin | | 分子名称: | 1,2-ETHANEDIOL, Jacalin-type lectin domain-containing protein | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2020-11-03 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Targeted disruption of pi-pi stacking in Malaysian banana lectin reduces mitogenicity while preserving antiviral activity.
Sci Rep, 11, 2021
|
|
7KG3
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | 著者 | Arvai, A, Brosey, C.A, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | 登録日 | 2020-10-15 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
4OUO
 
 | | anti-Bla g 1 scFv | | 分子名称: | CHLORIDE ION, SULFATE ION, anti Bla g 1 scFv | | 著者 | Mueller, G.A, Ankney, J.A, Glesner, J, Khurana, T, Edwards, L.L, Pedersen, L.C, Perera, L, Slater, J.E, Pomes, A, London, R.E. | | 登録日 | 2014-02-18 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization of an anti-Bla g 1 scFv: Epitope mapping and cross-reactivity.
Mol.Immunol., 59, 2014
|
|
4OXB
 
 | |
7KMV
 
 | | Structure of Malaysian Banana Lectin F84T | | 分子名称: | Jacalin-type lectin domain-containing protein, SULFATE ION | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2020-11-03 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Targeted disruption of pi-pi stacking in Malaysian banana lectin reduces mitogenicity while preserving antiviral activity.
Sci Rep, 11, 2021
|
|
3QIL
 
 | |
3QKE
 
 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter Salexigens complexed with Mg and D-Gluconate | | 分子名称: | D-gluconic acid, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | 著者 | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-02-01 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Cystal structure of D-mannonate dehydratase from Chromohalobacter salexigens complexed with Mg and D-Gluconate
To be Published
|
|
3QNM
 
 | | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function | | 分子名称: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | 著者 | Matthew, M.W, Ramagopal, U.A, Toro, R, Dickey, M, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2011-02-08 | | 公開日 | 2011-03-30 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function
To be Published
|
|
4O92
 
 | | Crystal structure of a Glutathione S-transferase from Pichia kudriavzevii (Issatchenkia orientalis), target EFI-501747 | | 分子名称: | Glutathione S-transferase, SULFATE ION | | 著者 | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Attonito, J.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-31 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of a Glutathione S-transferase from Pichia kudriavzevii (Issatchenkia orientalis), target EFI-501747
TO BE PUBLISHED
|
|
7L1U
 
 | | Orexin Receptor 2 (OX2R) in Complex with G Protein and Natural Peptide-Agonist Orexin B (OxB) | | 分子名称: | Engineered Guanine nucleotide-binding protein subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Hong, C, Byrne, N.J, Zamlynny, B, Tummala, S, Xiao, L, Shipman, J.M, Partridge, A.T, Minnick, C, Breslin, M.J, Rudd, M.T, Stachel, S.J, Rada, V.L, Kern, J.C, Armacost, K.A, Hollingsworth, S.A, O'Brien, J.A, Hall, D.L, McDonald, T.P, Strickland, C, Brooun, A, Soisson, S.M, Hollenstein, K. | | 登録日 | 2020-12-15 | | 公開日 | 2021-02-10 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Nat Commun, 12, 2021
|
|
7L1V
 
 | | Orexin Receptor 2 (OX2R) in Complex with G Protein and Small-Molecule Agonist Compound 1 | | 分子名称: | 4'-methoxy-N,N-dimethyl-3'-{[3-(2-{[2-(2H-1,2,3-triazol-2-yl)benzene-1-carbonyl]amino}ethyl)phenyl]sulfamoyl}[1,1'-biphenyl]-3-carboxamide, Engineered Guanine nucleotide-binding protein subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hong, C, Byrne, N.J, Zamlynny, B, Tummala, S, Xiao, L, Shipman, J.M, Partridge, A.T, Minnick, C, Breslin, M.J, Rudd, M.T, Stachel, S.J, Rada, V.L, Kern, J.C, Armacost, K.A, Hollingsworth, S.A, O'Brien, J.A, Hall, D.L, McDonald, T.P, Strickland, C, Brooun, A, Soisson, S.M, Hollenstein, K. | | 登録日 | 2020-12-15 | | 公開日 | 2021-02-10 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Nat Commun, 12, 2021
|
|
3QF0
 
 | | Crystal structure of the mutant T159V,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-01-20 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QFW
 
 | | Crystal structure of Rubisco-like protein from Rhodopseudomonas palustris | | 分子名称: | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, SULFATE ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2011-01-23 | | 公開日 | 2011-02-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.789 Å) | | 主引用文献 | Crystal structure of Rubisco-like protein from Rhodopseudomonas palustris
To be Published
|
|
4OVT
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM OCHROBACTERIUM ANTHROPI (Oant_3902), TARGET EFI-510153, WITH BOUND L-FUCONATE | | 分子名称: | 6-deoxy-L-galactonic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-14 | | 公開日 | 2014-01-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
7KXB
 
 | | Crystal structure of SARS-CoV-2 Nsp3 Macrodomain complex with PARG329 | | 分子名称: | BETA-MERCAPTOETHANOL, N-{3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]propyl}-N'-[2-(morpholin-4-yl)ethyl]thiourea, Non-structural protein 3, ... | | 著者 | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | 登録日 | 2020-12-03 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
4OWZ
 
 | |
3QMR
 
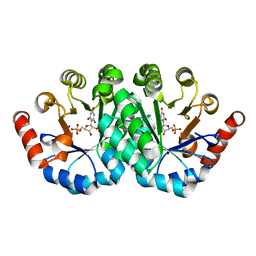 | | Crystal structure of the mutant R160A,V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-02-05 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.3213 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
7L12
 
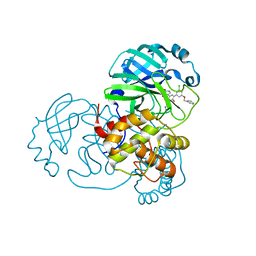 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 14 | | 分子名称: | (5S)-5-{3-[3-(benzyloxy)-5-chlorophenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2020-12-14 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L14
 
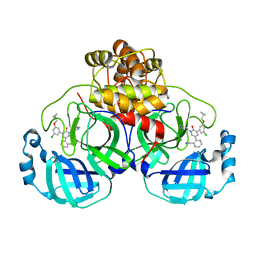 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 26 | | 分子名称: | 2-{3-[3-chloro-5-(cyclopropylmethoxy)phenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}benzonitrile, 3C-like proteinase | | 著者 | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2020-12-14 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L13
 
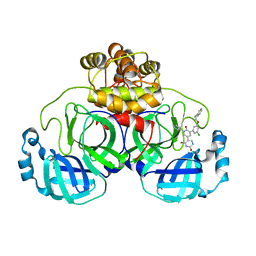 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 21 | | 分子名称: | (5S)-5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]phenyl}-2-oxo[2H-[1,3'-bipyridine]]-5-yl)pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2020-12-14 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
