6FFH
 
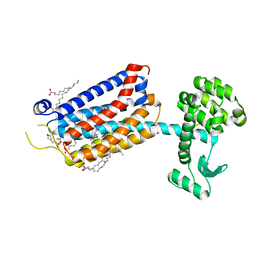 | | Crystal Structure of mGluR5 in complex with Fenobam at 2.65 A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(3-chlorophenyl)-3-(3-methyl-5-oxidanylidene-4~{H}-imidazol-2-yl)urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
1DF1
 
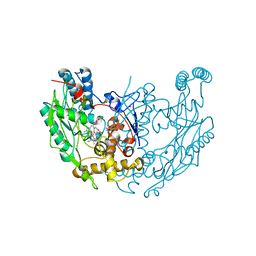 | | MURINE INOSOXY DIMER WITH ISOTHIOUREA BOUND IN THE ACTIVE SITE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYLISOTHIOUREA, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Rosenfeld, R.J, Arvai, A.S, Ghosh, D.K, Ghosh, S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-16 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization.
EMBO J., 18, 1999
|
|
4WK5
 
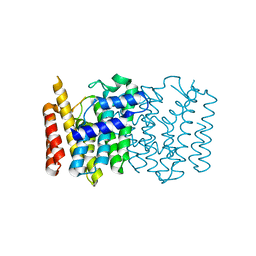 | | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458 | | Descriptor: | Geranyltranstransferase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458
To be published
|
|
6FCR
 
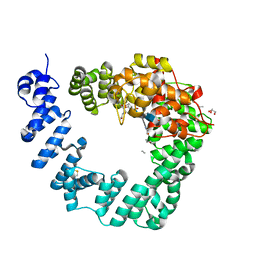 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMtetrapeptide-NAG-anhNAMtetrapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1TW6
 
 | | Structure of an ML-IAP/XIAP chimera bound to a 9mer peptide derived from Smac | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Vucic, D, Wallweber, H.J.A, Das, K, Shin, H, Elliott, L.O, Kadkhodayan, S, Deshayes, K, Salvesen, G.S, Fairbrother, W.J. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Engineering ML-IAP to produce an extraordinarily potent caspase 9 inhibitor: implications for Smac-dependent anti-apoptotic activity of ML-IAP
Biochem.J., 385, 2005
|
|
6FCS
 
 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMpentapeptide-NAG-NAMpentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4WMJ
 
 | |
1DCP
 
 | | DCOH, A BIFUNCTIONAL PROTEIN-BINDING TRANSCRIPTIONAL COACTIVATOR, COMPLEXED WITH BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, DCOH | | Authors: | Cronk, J.D, Endrizzi, J.A, Alber, T. | | Deposit date: | 1996-05-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution structures of the bifunctional enzyme and transcriptional coactivator DCoH and its complex with a product analogue.
Protein Sci., 5, 1996
|
|
6FBT
 
 | | The X-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa in complex with the reaction product NAG-anhNAMpentapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, NAG-anhNAMpentapeptide, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FC8
 
 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-(3-fluorophenyl)-4-[[(3~{S})-piperidin-3-yl]amino]thieno[3,2-c]pyridine-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
4WG2
 
 | | P411BM3-CIS T438S I263F regioselective C-H amination catalyst | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hyster, T.K, Farwell, C.C, Buller, A.R, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2014-09-17 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Enzyme-controlled nitrogen-atom transfer enables regiodivergent C-h amination.
J.Am.Chem.Soc., 136, 2014
|
|
6FCK
 
 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-phenyl-4-[[(3~{S})-piperidin-3-yl]amino]-1~{H}-indole-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FFI
 
 | | Crystal Structure of mGluR5 in complex with MMPEP at 2.2 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-(3-methoxyphenyl)ethynyl]-6-methyl-pyridine, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
1DKS
 
 | | CKSHS1: HUMAN CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1 IN COMPLEX WITH PHOSPHATE | | Descriptor: | CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1, PHOSPHATE ION | | Authors: | Bourne, Y, Arvai, A.S, Tainer, J.A. | | Deposit date: | 1995-11-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain.
J.Mol.Biol., 249, 1995
|
|
4WN9
 
 | | Structure of the Nitrogenase MoFe Protein from Clostridium pasteurianum Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
4WWQ
 
 | | Apo structure of the Grb7 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 7, MALONIC ACID | | Authors: | Watson, G.M, Ambaye, N.D, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
6FNP
 
 | | Crystal structure of ECF-CbrT, a cobalamin transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Santos, J.A, Rempel, S, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Functional and structural characterization of an ECF-type ABC transporter for vitamin B12.
Elife, 7, 2018
|
|
4WUT
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
4X6S
 
 | | Grb7 SH2 domain with phosphotyrosine mimetic inhibitor peptide | | Descriptor: | Growth factor receptor-bound protein 7, Phosphotyrosine mimetic inhibitor peptide G7-TEM1 | | Authors: | Watson, G.M, Panjikar, S, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-09-23 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
4X09
 
 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
1E4I
 
 | | 2-deoxy-2-fluoro-beta-D-glucosyl/enzyme intermediate complex of the beta-glucosidase from Bacillus polymyxa | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE | | Authors: | Sanz-Aparicio, J, Gonzalez, B, Hermoso, J.A, Arribas, J.C, Canada, F.J, Polaina, J. | | Deposit date: | 2000-07-06 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Increased Resistance to Thermal Denaturation Induced by Single Amino Acid Substitution in the Sequence of Beta-Glucosidase a from Bacillus Polymyxa.
Proteins: Struct.,Funct., Genet., 33, 1998
|
|
4WT7
 
 | | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol | | Descriptor: | ABC transporter substrate binding protein (Ribose), CHLORIDE ION, D-allitol | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol
To be published
|
|
4WNA
 
 | | Structure of the Nitrogenase MoFe Protein from Azotobacter vinelandii Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Zhang, L, Einsle, O, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
4WS9
 
 | | Crystal structure of sMAT N159G from Sulfolobus solfataricus | | Descriptor: | PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Wang, F, Brady, E.L, Singh, S, Clinger, J.A, Huber, T.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of sMAT N159G from Sulfolobus solfataricus.
To Be Published
|
|
6FTD
 
 | | Deinococcus radiodurans BphP PAS-GAF H290T mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, ... | | Authors: | Edlund, P, Takala, H, Westenhoff, S, Ihalainen, J.A. | | Deposit date: | 2018-02-21 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Coordination of the biliverdin D-ring in bacteriophytochromes.
Phys Chem Chem Phys, 20, 2018
|
|
