4MHV
 
 | | Crystal structure of the PNT domain of human ETS2 | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Newman, J.A, Cooper, C.D.O, Krojer, T, Shrestha, L, Burgess-brown, N, Arrowsmith, C.H, Bountra, C, Edwards, A, Gileadi, O. | | 登録日 | 2013-08-30 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of the PNT domain of human ETS2
To be Published
|
|
4MF5
 
 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound | | 分子名称: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | 著者 | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-08-27 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound
To be Published
|
|
4NHB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio desulfuricans (Ddes_1525), Target EFI-510107, with bound sn-glycerol-3-phosphate | | 分子名称: | IODIDE ION, SN-GLYCEROL-3-PHOSPHATE, TRAP dicarboxylate transporter-DctP subunit | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-11-04 | | 公開日 | 2013-11-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3KPC
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | 著者 | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | 登録日 | 2009-11-16 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
4NF0
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOMONAS AERUGINOSA PAO1 (PA4616), TARGET EFI-510182, WITH BOUND L-Malate | | 分子名称: | (2S)-2-hydroxybutanedioic acid, Probable c4-dicarboxylate-binding protein, SULFATE ION | | 著者 | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-10-30 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NG7
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Citrobacter koseri (CKO_04899), Target EFI-510094, apo, open structure | | 分子名称: | TRAP periplasmic solute binding protein | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-11-01 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NQA
 
 | | Crystal structure of liganded hRXR-alpha/hLXR-beta heterodimer on DNA | | 分子名称: | (9cis)-retinoic acid, 5'-D(*TP*AP*AP*GP*GP*TP*CP*AP*CP*TP*TP*CP*AP*GP*GP*TP*CP*A)-3', 5'-D(*TP*AP*TP*GP*AP*CP*CP*TP*GP*AP*AP*GP*TP*GP*AP*CP*CP*T)-3', ... | | 著者 | Lou, X.H, Toresson, G, Benod, C, Suh, J.H, Phillips, K.J, Webb, P, Gustafsson, J.A. | | 登録日 | 2013-11-24 | | 公開日 | 2014-02-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structure of the retinoid X receptor alpha-liver X receptor beta (RXR alpha-LXR beta ) heterodimer on DNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1ZZQ
 
 | | Rat nNOS D597N mutant with L-N(omega)-Nitroarginine-(4R)-amino-L-proline amide bound | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | 著者 | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | 登録日 | 2005-06-14 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
6I1S
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with FKBP12 and the inhibitor E6201 | | 分子名称: | (4~{S},5~{R},6~{Z},9~{S},10~{S},12~{E})-16-(ethylamino)-4,5-dimethyl-9,10,18-tris(oxidanyl)-3-oxabicyclo[12.4.0]octadeca-1(14),6,12,15,17-pentaene-2,8-dione, 1,2-ETHANEDIOL, Activin receptor type-1, ... | | 著者 | Williams, E.P, Pinkas, D.M, Fortin, J, Newman, J.A, Bradshaw, W.J, Mahajan, P, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | 登録日 | 2018-10-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas.
Cancer Cell, 37, 2020
|
|
4NN3
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens (Desal_2161), Target EFI-510109, with bound orotic acid | | 分子名称: | CHLORIDE ION, OROTIC ACID, TRAP dicarboxylate transporter, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-11-16 | | 公開日 | 2013-12-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NQ8
 
 | | Crystal structure of a trap periplasmic solute binding protein from Bordetella bronchispeptica (bb3421), target EFI-510039, with density modeled as pantoate | | 分子名称: | CHLORIDE ION, PANTOATE, Putative periplasmic substrate-binding transport protein, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-11-24 | | 公開日 | 2014-01-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3KPD
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine. | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | 著者 | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | 登録日 | 2009-11-16 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
4NO7
 
 | | Human Glucokinase in complex with a nanomolar activator. | | 分子名称: | (2R)-2-[3-chloro-4-(methylsulfonyl)phenyl]-3-[(1R)-3-oxocyclopentyl]-N-(pyrazin-2-yl)propanamide, Glucokinase, alpha-D-glucopyranose | | 著者 | Petit, P, Ferry, G, Antoine, M, Boutin, J.A, Kotschy, A, Perron-Sierra, F, Mamelli, L, Vuillard, L. | | 登録日 | 2013-11-19 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The active conformation of human glucokinase is not altered by allosteric activators.
Acta Crystallogr. D Biol. Crystallogr., 67, 2011
|
|
4NX5
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | 分子名称: | 1,2-ETHANEDIOL, 6-AZA URIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | 登録日 | 2013-12-08 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.591 Å) | | 主引用文献 | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate
To be Published
|
|
4NY9
 
 | | Crystal Structure Of the Human PXR-LBD In Complex With N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-3-hydroxy-3-methylbutanamide | | 分子名称: | GLYCEROL, N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-3-hydroxy-3-methylbutanamide, Nuclear receptor subfamily 1 group I member 2 | | 著者 | Khan, J.A, Camac, D.M. | | 登録日 | 2013-12-10 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of the CCR1 antagonist, BMS-817399, for the treatment of rheumatoid arthritis.
J.Med.Chem., 57, 2014
|
|
4NX1
 
 | | Crystal structure of a trap periplasmic solute binding protein from Sulfitobacter sp. nas-14.1, target EFI-510292, with bound alpha-D-taluronate | | 分子名称: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-talopyranuronic acid | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-08 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O11
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-hydroxyuridine 5'-monophosphate | | 分子名称: | 1,2-ETHANEDIOL, 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2013-12-14 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-hydroxyuridine 5'-monophosphate
To be Published
|
|
4NQL
 
 | | The crystal structure of the DUB domain of AMSH orthologue, Sst2 from S. pombe, in complex with lysine 63-linked diubiquitin | | 分子名称: | 1,2-ETHANEDIOL, AMSH-like protease sst2, Ubiquitin, ... | | 著者 | Ronau, J.A, Shrestha, R.K, Das, C. | | 登録日 | 2013-11-25 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Insights into the mechanism of deubiquitination by JAMM deubiquitinases from cocrystal structures of the enzyme with the substrate and product.
Biochemistry, 53, 2014
|
|
4OVQ
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS, TARGET EFI-510230, WITH BOUND BETA-D-GLUCURONATE | | 分子名称: | CHLORIDE ION, TRAP dicarboxylate ABC transporter, substrate-binding protein, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-11 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1, TARGET EFI-510292, WITH BOUND ALPHA-D-MANURONATE | | 分子名称: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-mannopyranuronic acid | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-11 | | 公開日 | 2014-01-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O8M
 
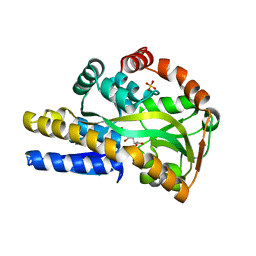 | | Crystal structure of a trap periplasmic solute binding protein actinobacillus succinogenes 130z, target EFI-510004, with bound L-galactonate | | 分子名称: | CHLORIDE ION, L-galactonic acid, SULFATE ION, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-28 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OGC
 
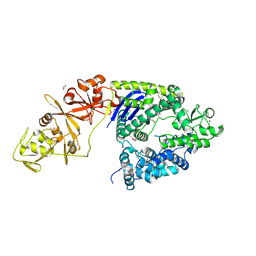 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | 分子名称: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | 著者 | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | 登録日 | 2014-01-15 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
1ZZU
 
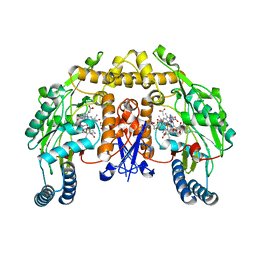 | | Rat nNOS D597N/M336V double mutant with L-N(omega)-Nitroarginine-2,4-L-Diaminobutyric Amide Bound | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | 著者 | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | 登録日 | 2005-06-14 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
4OUO
 
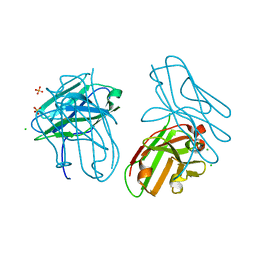 | | anti-Bla g 1 scFv | | 分子名称: | CHLORIDE ION, SULFATE ION, anti Bla g 1 scFv | | 著者 | Mueller, G.A, Ankney, J.A, Glesner, J, Khurana, T, Edwards, L.L, Pedersen, L.C, Perera, L, Slater, J.E, Pomes, A, London, R.E. | | 登録日 | 2014-02-18 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization of an anti-Bla g 1 scFv: Epitope mapping and cross-reactivity.
Mol.Immunol., 59, 2014
|
|
4OXB
 
 | |
