5VFC
 
 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
5VFX
 
 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
5VGS
 
 | | Crystal structure of lachrymatory factor synthase from Allium cepa in complex with crotyl alcohol | | Descriptor: | (2E)-but-2-en-1-ol, (2Z)-but-2-en-1-ol, Lachrymatory-factor synthase | | Authors: | Silvaroli, J.A, Pleshinger, M.J, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzyme That Makes You Cry-Crystal Structure of Lachrymatory Factor Synthase from Allium cepa.
ACS Chem. Biol., 12, 2017
|
|
5VMQ
 
 | |
1L30
 
 | |
1L32
 
 | |
1L25
 
 | |
1L31
 
 | |
1L35
 
 | |
1L28
 
 | |
1L27
 
 | |
4ZVU
 
 | |
4ZVQ
 
 | |
1L29
 
 | |
1L26
 
 | |
1A1R
 
 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | Descriptor: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | Authors: | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
6YI9
 
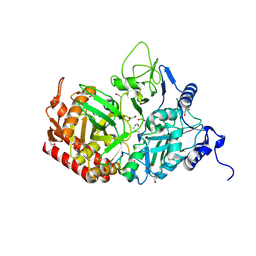 | | Crystal structure of the rat cytosolic PCK1, acetylated on Lys244 | | Descriptor: | 1,2-ETHANEDIOL, Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | | Authors: | Latorre-Muro, P, Baeza, J, Hurtado-Guerrero, R, Hicks, T, Delso, I, Hernandez-Ruiz, C, Velazquez-Campoy, A, Lawton, A.J, Angulo, J, Denu, J.M, Carrodeguas, J.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Self-acetylation at the active site of phosphoenolpyruvate carboxykinase (PCK1) controls enzyme activity.
J.Biol.Chem., 296, 2021
|
|
7B2X
 
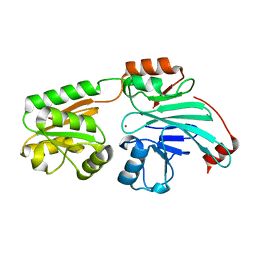 | | Crystal structure of human 5' exonuclease Appollo H61Y variant | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
2M86
 
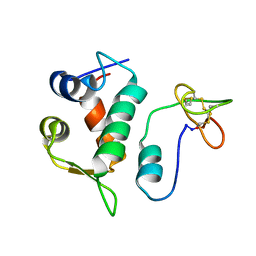 | | Solution structure of Hdm2 with engineered cyclotide | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, MCo-PMI | | Authors: | Majumder, S, Ji, Y, Millard, M, Borra, R, Bi, T, Elnagar, A.Y, Neamati, N, Camarero, J.A. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | In Vivo Activation of the p53 Tumor Suppressor Pathway by an Engineered Cyclotide.
J.Am.Chem.Soc., 135, 2013
|
|
2M19
 
 | | Solution structure of the Haloferax volcanii HVO 2177 protein | | Descriptor: | Molybdopterin converting factor subunit 1 | | Authors: | Li, Y, Maciejewski, M.W, Martin, J, Jin, K, Zhang, Y, Lu, M, Maupin-Furlow, J.A, Hao, B. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
2MAP
 
 | |
4W2O
 
 | |
4W2P
 
 | | Anti-Marburgvirus Nucleoprotein Single Domain Antibody C | | Descriptor: | ACETATE ION, Anti-Marburgvirus Nucleoprotein Single Domain Antibody C, SODIUM ION | | Authors: | Taylor, A.B, Garza, J.A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Unveiling a Drift Resistant Cryptotope withinMarburgvirusNucleoprotein Recognized by Llama Single-Domain Antibodies.
Front Immunol, 8, 2017
|
|
4W2Q
 
 | |
