5PHM
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09455a | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, N-OXALYLGLYCINE, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6CRV
 
 | | SARS Spike Glycoprotein, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
5PNO
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 218) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5POE
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10188a and N07807b | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4-phenyl-5-(1H-pyrrol-1-yl)-1H-pyrazole, 6-amino-1-methylquinolin-2(1H)-one, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.518 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PI1
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 14) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PIG
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 29) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3FIF
 
 | | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A. | | Descriptor: | Uncharacterized ligand, Uncharacterized lipoprotein ygdR | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-11 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
5PIX
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 46) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3FIW
 
 | | Structure of SCO0253, a Tetr-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative tetR-family transcriptional regulator | | Authors: | Singer, A.U, Xu, X, Chang, C, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SCO0253, a Tetr-family transcriptional regulator from Streptomyces coelicolor
To be Published
|
|
5POQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10974a | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(methylsulfonyl)piperazin-1-yl]ethan-1-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3FJK
 
 | |
5PP1
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 1) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PJC
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 61) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PJT
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 78) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PK8
 
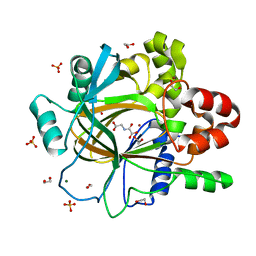 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 93) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6HS4
 
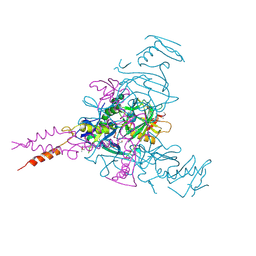 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118 | | Descriptor: | 1-[2-[3-oxidanyl-4-(4-phenyl-1~{H}-pyrazol-5-yl)phenoxy]ethyl]piperidine-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Stuart, D.I. | | Deposit date: | 2018-09-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
5PQ8
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 45) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3EYE
 
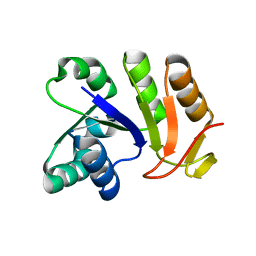 | | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli | | Descriptor: | PTS system N-acetylgalactosamine-specific IIB component 1 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli
To be Published
|
|
5PG5
 
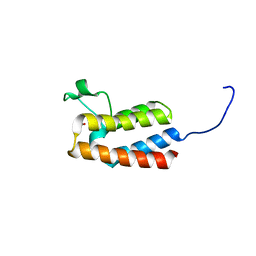 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 169) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PKO
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 109) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3HNQ
 
 | | Crystal Structure of Virulence protein STM3117 from Salmonella typhimurium. Northeast Structural Genomics Consortium target id StR274 | | Descriptor: | Virulence protein STM3117 | | Authors: | Seetharaman, J, Su, M, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-31 | | Release date: | 2009-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Virulence protein STM3117 from Salmonella typhimurium. Northeast Structural Genomics Consortium target id StR274
To be Published
|
|
5PL4
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 125) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4BNF
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2- phenoxy-5-propylphenol | | Descriptor: | 2-phenoxy-5-propyl-phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
5PLL
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 142) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3FK3
 
 | | Structure of the Yeats Domain, Yaf9 | | Descriptor: | Protein AF-9 homolog | | Authors: | Wang, A.Y, Schulze, J.M, Skordalakes, E, Berger, J.M, Rine, J, Kobor, M.S. | | Deposit date: | 2008-12-15 | | Release date: | 2009-10-27 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
