5UTU
 
 | | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD
To Be Published
|
|
4YUI
 
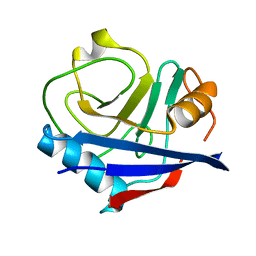 | | Multiconformer synchrotron model of CypA at 180 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
8EAP
 
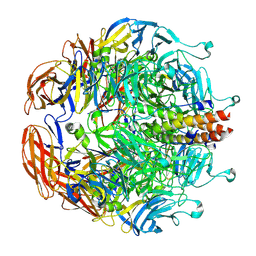 | |
8EB7
 
 | |
5UOH
 
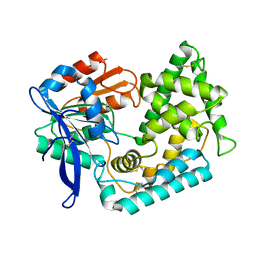 | | Crystal Structure of Hip1 (Rv2224c) T466A mutant | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Baikovitz, J, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
1KDN
 
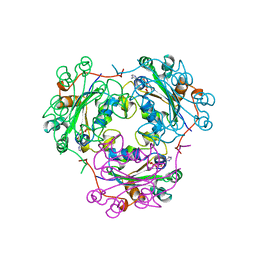 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cherfils, J, Xu, Y.W, Morera, S, Janin, J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
4YM5
 
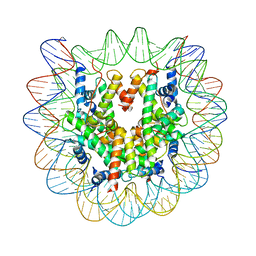 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
8EAN
 
 | |
8EDO
 
 | |
1TRB
 
 | |
4DNW
 
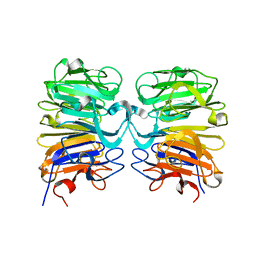 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
8EAO
 
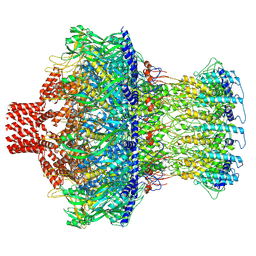 | |
5LM4
 
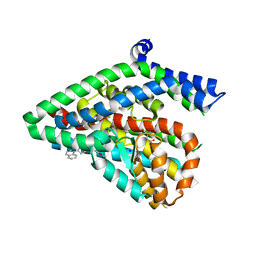 | | Structure of the thermostalilized EAAT1 cryst-II mutant in complex with L-ASP and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
1YAT
 
 | |
1KLM
 
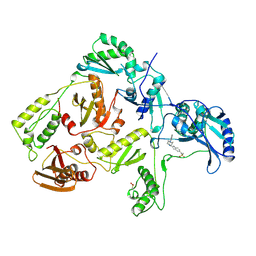 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH BHAP U-90152 | | Descriptor: | (1-(5-METHANSULPHONAMIDO-1H-INDOL-2-YL-CARBONYL)4-[METHYLAMINO)PYRIDINYL]PIPERAZINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R.M, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1997-03-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
4YND
 
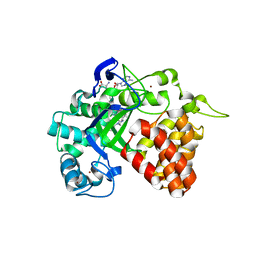 | | The Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2 | | Descriptor: | N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[(2R)-2-hydroxy-2-(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Sweis, R.F, Wang, Z, Algire, M, Arrowsmith, C.H, Brown, P.J, Chiang, G.C, Guo, J, Jakob, C.G, Kennedy, S, Li, F, Soni, N.B, Vedadi, M, Pappano, W.N. | | Deposit date: | 2015-03-09 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2.
Acs Med.Chem.Lett., 6, 2015
|
|
8EDN
 
 | |
1TUI
 
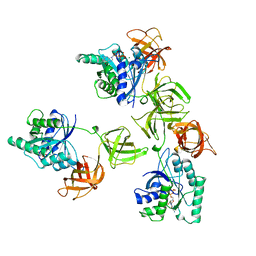 | | INTACT ELONGATION FACTOR TU IN COMPLEX WITH GDP | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Polekhina, G, Thirup, S, Kjeldgaard, M, Nissen, P, Lippmann, C, Nyborg, J. | | Deposit date: | 1996-05-23 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unwinding in the effector region of elongation factor EF-Tu-GDP.
Structure, 4, 1996
|
|
1O6E
 
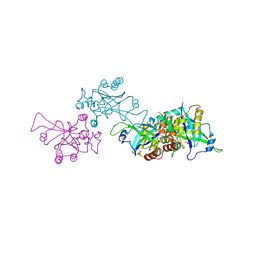 | | Epstein-Barr virus protease | | Descriptor: | CAPSID PROTEIN P40, PHOSPHORYLISOPROPANE | | Authors: | Buisson, M, Hernandez, J, Lascoux, D, Schoehn, G, Forest, E, Arlaud, G, Seigneurin, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2002-09-13 | | Release date: | 2002-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Epstein-Barr Virus Protease Shows Rearrangement of the Processed C Terminus
J.Mol.Biol., 324, 2002
|
|
8EDL
 
 | |
1TZE
 
 | |
1UCW
 
 | |
1O8N
 
 | | The active site of the molybdenum cofactor biosynthetic protein domain Cnx1G | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN | | Authors: | Kuper, J, Winking, J, Hecht, H.J, Schwarz, G, Mendel, R.R. | | Deposit date: | 2002-11-28 | | Release date: | 2003-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G
Arch.Biochem.Biophys., 411, 2003
|
|
8E4G
 
 | |
8EB3
 
 | |
