5XUP
 
 | | Crystal structure of TRF1 and TERB1 | | Descriptor: | Telomere repeats-binding bouquet formation protein 1, Telomeric repeat-binding factor 1 | | Authors: | Long, J, Huang, C, Wu, J, Lei, M. | | Deposit date: | 2017-06-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Telomeric TERB1-TRF1 interaction is crucial for male meiosis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6L90
 
 | | Crystal structure of ugt transferase enzyme | | Descriptor: | Glycosyltransferase, SULFATE ION | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
5XVC
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
1VID
 
 | | CATECHOL O-METHYLTRANSFERASE | | Descriptor: | 3,5-DINITROCATECHOL, CATECHOL O-METHYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Vidgren, J, Svensson, L.A, Liljas, A. | | Deposit date: | 1996-01-05 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of catechol O-methyltransferase.
Nature, 368, 1994
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
6O57
 
 | |
6LEB
 
 | | Staphylococcus aureus surface protein SdrC mutant-P366H | | Descriptor: | GLYCEROL, MAGNESIUM ION, Ser-Asp rich fibrinogen-binding, ... | | Authors: | Hang, T, Zhang, M, Wang, J. | | Deposit date: | 2019-11-25 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the intermolecular interaction of the adhesin SdrC in the pathogenicity of Staphylococcus aureus.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3CFH
 
 | |
6O6P
 
 | | Structure of the regulator FasR from Mycobacterium tuberculosis in complex with DNA | | Descriptor: | DNA-forward, DNA-reverse, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.851 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
2J7T
 
 | | Crystal structure of human serine threonine kinase-10 bound to SU11274 | | Descriptor: | (3Z)-N-(3-CHLOROPHENYL)-3-({3,5-DIMETHYL-4-[(4-METHYLPIPERAZIN-1-YL)CARBONYL]-1H-PYRROL-2-YL}METHYLENE)-N-METHYL-2-OXOINDOLINE-5-SULFONAMIDE, ACETATE ION, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Das, S, Debreczeni, J, Sobott, F, Watt, S, Savitsky, P, Eswaran, J, Turnbull, A.P, Papagrigoriou, E, Ugochukwa, E, Gorrec, F, Umeano, C.C, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
4M8X
 
 | | GS-8374, a Novel Phosphonate-Containing Inhibitor of HIV-1 Protease, Effectively Inhibits HIV PR Mutants with Amino Acid Insertions | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Grantz saskova, K, Brynda, J, Rezacova, P, Kozisek, M, Konvalinka, J. | | Deposit date: | 2013-08-14 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GS-8374, a prototype phosphonate-containing inhibitor of HIV-1 protease, effectively inhibits protease mutants with amino acid insertions.
J.Virol., 88, 2014
|
|
6OC5
 
 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
2O1M
 
 | | Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572 | | Descriptor: | Probable amino-acid ABC transporter extracellular-binding protein ytmK, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572.
To be Published
|
|
6OL2
 
 | | Crystallography of novel WNK1 and WNK3 inhibitors discovered from high-throughput-screening | | Descriptor: | ACETATE ION, GLYCEROL, N-[2-(5,8-dimethoxy-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]-2-iodobenzamide, ... | | Authors: | Chlebowicz, J, Akella, R, Sekulski, K, Humphreys, J.M, Durbacz, M.Z, He, H, Rodan, A, Posner, B, Goldsmith, E.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallography of novel WNK1 and WNK3 inhibitors discovered from high throughput screening
To Be Published
|
|
8PP7
 
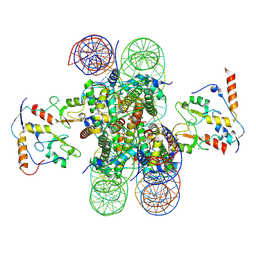 | | human RYBP-PRC1 bound to mononucleosome | | Descriptor: | DNA (215-mer), E3 ubiquitin-protein ligase RING2, Histone H2A, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6OSQ
 
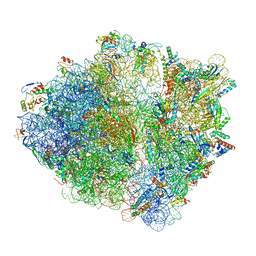 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
8ACM
 
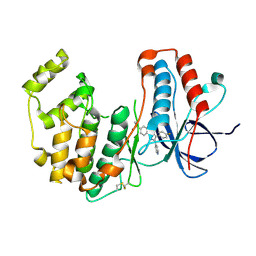 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACO
 
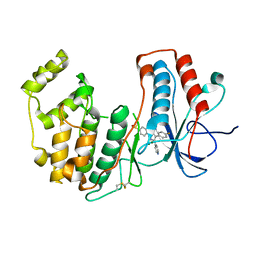 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
5APG
 
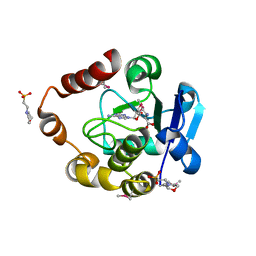 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
2YGT
 
 | | Clostridium perfringens delta-toxin | | Descriptor: | DELTA TOXIN, GLYCEROL, IMIDAZOLE, ... | | Authors: | Huyet, J, Naylor, C.E, Gibert, M, Popoff, M.R, Basak, A.K. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Clostridium Perfringens Delta Toxin Pore Formation.
Plos One, 8, 2013
|
|
6L3U
 
 | | Human Cx31.3/GJC3 connexin hemichannel in the presence of calcium | | Descriptor: | CALCIUM ION, Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
2NVV
 
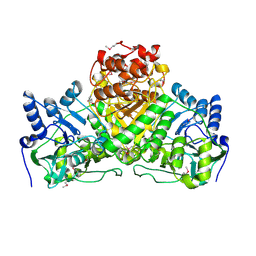 | | Crystal Structure of the Putative Acetyl-CoA hydrolase/transferase PG1013 from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR16. | | Descriptor: | Acetyl-CoA hydrolase/transferase family protein, ZINC ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Yong, W, Ho, C.K, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Acetyl-CoA hydrolase/transferase PG1013 from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR16
To be Published
|
|
2GAO
 
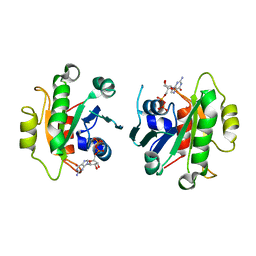 | | Crystal Structure of Human SAR1a in Complex With GDP | | Descriptor: | GTP-binding protein SAR1a, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Wang, J, Dimov, S, Tempel, W, Yaniw, D, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human SAR1a in Complex With GDP
To be Published
|
|
8I80
 
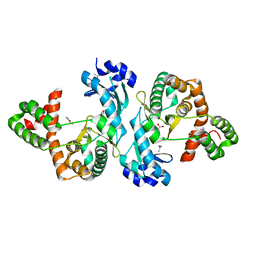 | | Crystal structure of Cph001-D189N | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J. | | Deposit date: | 2023-02-02 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I86
 
 | | Crystal structure of Cph001-D189N in complex with GTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
