3FN8
 
 | |
6C3O
 
 | | Cryo-EM structure of human KATP bound to ATP and ADP in quatrefoil form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Lee, K.P.K, Chen, J, MacKinnon, R. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of human KATP in complex with ATP and ADP
Elife, 6, 2017
|
|
6E0E
 
 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
6E12
 
 | | Crystal Structure of the Alr8543 protein in complex with Oleic Acid and magnesium ion from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR141 | | Descriptor: | Alr8543 protein, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Xiao, R, Ciccosanti, C, Patel, D, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-07-09 | | Release date: | 2018-07-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Alr8543 protein in complex with Oleic Acid and magnesium ion from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR141
To Be Published
|
|
6C5V
 
 | | An anti-gH/gL antibody that neutralizes dual-tropic infection defines a site of vulnerability on Epstein-Barr virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab AMMO1 heavy chain, Antibody Fab AMMO1 light chain, ... | | Authors: | Snijder, J, Ortego, M.S, Weidle, C, Stuart, A.B, Gray, M.A, McElrath, M.J, Pancera, M, Veesler, D, McGuire, A.T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-01-16 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus.
Immunity, 48, 2018
|
|
7ZU0
 
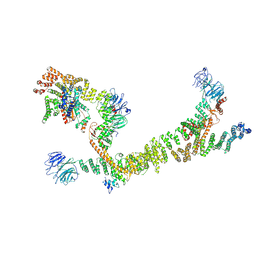 | | HOPS tethering complex from yeast | | Descriptor: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar morphogenesis protein 6, ... | | Authors: | Shvarev, D, Schoppe, J, Koenig, C, Perz, A, Fuellbrunn, N, Kiontke, S, Langemeyer, L, Januliene, D, Schnelle, K, Kuemmel, D, Froehlich, F, Moeller, A, Ungermann, C. | | Deposit date: | 2022-05-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the endosomal CORVET tethering complex.
Nat Commun, 15, 2024
|
|
6C9W
 
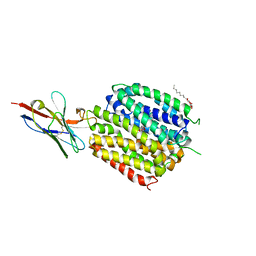 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1FG0
 
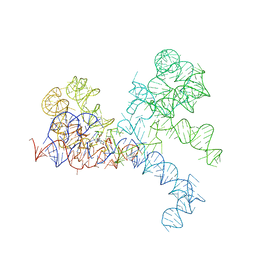 | | LARGE RIBOSOMAL SUBUNIT COMPLEXED WITH A 13 BP MINIHELIX-PUROMYCIN COMPOUND | | Descriptor: | 23S RIBOSOMAL RNA, 5'-R(CCGGCGGGCUGGUUCAAACCGGCCCGCCGGACC)-3'-5'-R(P-PUROMYCIN)-3' | | Authors: | Nissen, P, Hansen, J, Ban, N, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-26 | | Release date: | 2000-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of ribosome activity in peptide bond synthesis.
Science, 289, 2000
|
|
6CHX
 
 | |
3FZE
 
 | | Structure of the 'minimal scaffold' (ms) domain of Ste5 that cocatalyzes Fus3 phosphorylation by Ste7 | | Descriptor: | Protein STE5 | | Authors: | Good, M.C, Tang, G, Singleton, J, Remenyi, A, Lim, W.A. | | Deposit date: | 2009-01-25 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation.
Cell(Cambridge,Mass.), 136, 2009
|
|
3FQ9
 
 | | Design of an insulin analog with enhanced receptor-binding selectivity. Rationale, structure, and therapeutic implications | | Descriptor: | Insulin, ZINC ION | | Authors: | Zhao, M, Wan, Z.L, Whittaker, L, Xu, B, Phillips, N, Katsoyannis, P, Whittaker, J, Weiss, M.A. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of an insulin analog with enhanced receptor binding selectivity: rationale, structure, and therapeutic implications.
J.Biol.Chem., 284, 2009
|
|
3FVS
 
 | | Human Kynurenine Aminotransferase I in complex with Glycerol | | Descriptor: | GLYCEROL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
6CL4
 
 | | LipC12 - Lipase from metagenomics | | Descriptor: | Lipase C12 | | Authors: | Iulek, J, Martini, V.P, Krieger, N, Glogauer, A, Souza, E.M. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure solution and analyses of the first true lipase obtained from metagenomics indicate potential for increased thermostability.
N Biotechnol, 53, 2019
|
|
6CLF
 
 | | 1.15 A MicroED structure of GSNQNNF at 1.9 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
3FQV
 
 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-[3-(3-methoxy-4-phenylphenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3S)-3-(2-methoxybiphenyl-4-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2009-01-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.
J.Mol.Biol., 387, 2009
|
|
6CLO
 
 | | 1.15 A MicroED structure of GSNQNNF at 2.1 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
3FR2
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 9-benzyl-2,3,4,9-tetrahydro-1H-carbazole-8-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Rondahl, L, Uppenberg, J, Svensson, S, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2HEB
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
3FRT
 
 | |
3FR5
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 5-(3-carbamoylbenzyl)-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-4-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Rondahl, L, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FRN
 
 | | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima MSB8 | | Descriptor: | Flagellar protein FlgA, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Hu, S, Bain, K, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima
To be Published
|
|
2HEA
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
2HEF
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
6CPL
 
 | | Crystal structure of DR11 presenting the gag293 epitope | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
3DFQ
 
 | |
