5JIG
 
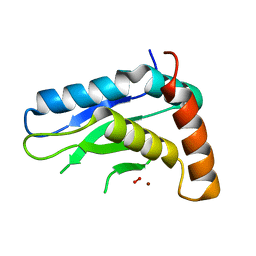 | | Crytsal structure of Wss1 from S. pombe | | Descriptor: | NICKEL (II) ION, OXYGEN MOLECULE, Ubiquitin and WLM domain-containing metalloprotease SPCC1442.07c | | Authors: | Groll, M, Stingele, J, Boulton, S. | | Deposit date: | 2016-04-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN.
Mol.Cell, 64, 2016
|
|
1H8O
 
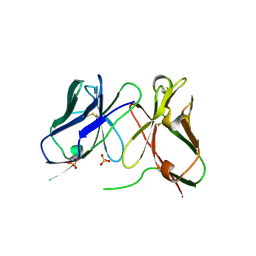 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment. | | Descriptor: | MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
8BSD
 
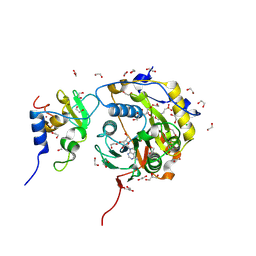 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Oberthuer, D, Sprenger, J. | | Deposit date: | 2022-11-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Tubercidin bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
To Be Published
|
|
8B8R
 
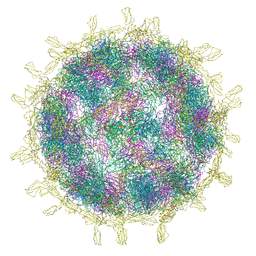 | | Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55) | | Descriptor: | DECAY ACCELERATING FACTOR (CD55), SPHINGOSINE, VP1, ... | | Authors: | Stuart, D.I, Ren, J, Zhou, D, Qin, L. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
3BZ5
 
 | | Functional domain of InlJ from Listeria monocytogenes includes a cysteine ladder | | Descriptor: | CHLORIDE ION, Internalin-J, SULFATE ION | | Authors: | Bublitz, M, Holland, C, Sabet, C, Reichelt, J, Cossart, P, Heinz, D.W, Bierne, H, Schubert, W.D. | | Deposit date: | 2008-01-17 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and standardized geometric analysis of InlJ, a listerial virulence factor and leucine-rich repeat protein with a novel cysteine ladder.
J.Mol.Biol., 378, 2008
|
|
5BYI
 
 | |
1R52
 
 | | Crystal structure of the bifunctional chorismate synthase from Saccharomyces cerevisiae | | Descriptor: | Chorismate synthase | | Authors: | Quevillon-Cheruel, S, Leulliot, N, Meyer, P, Graille, M, Bremang, M, Blondeau, K, Sorel, I, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-10-09 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of the bifunctional chorismate synthase from Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
1R8H
 
 | | Comparison of the structure and DNA binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus | | Descriptor: | PHOSPHATE ION, Regulatory protein E2 | | Authors: | Dell, G, Wilkinson, K.W, Tranter, R, Parish, J, Brady, R.L, Gaston, K. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of the structure and DNA-binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus.
J.Mol.Biol., 334, 2003
|
|
8BW4
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-4-(3-fluoranylthiophen-2-yl)carbonyl-N-(4-methoxyphenyl)-2-methyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
1H29
 
 | | Sulfate respiration in Desulfovibrio vulgaris Hildenborough: Structure of the 16-heme Cytochrome c HmcA at 2.5 A resolution and a view of its role in transmembrane electron transfer | | Descriptor: | HEME C, HIGH-MOLECULAR-WEIGHT CYTOCHROME C | | Authors: | Matias, P.M, Coelho, A.V, Valente, F.M.A, Placido, D, Legall, J, Xavier, A.V, Pereira, I.A.C, Carrondo, M.A. | | Deposit date: | 2002-08-01 | | Release date: | 2002-10-02 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Sulfate Respiration in Desulfovibrio Vulgaris Hildenborough: Structure of the 16-Heme Cytochrome C Hmca at 2.5 A Resolution and a View of its Role in Transmembrane Electron Transfer
J.Biol.Chem., 277, 2002
|
|
5JN6
 
 | |
5C04
 
 | |
3LGU
 
 | | Y162A mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
8BW3
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2S)-N-(cyclopropylmethyl)-2-methyl-4-(1-methyl-1H-pyrrole-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BET
 
 | |
3BUY
 
 | | MHC-I in complex with peptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Rossjohn, J, La Gruta, N.L, Purcell, A.W, Turner, S.J, Dunstone, M.A. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epitope-specific TCRbeta repertoire diversity imparts no functional advantage on the CD8+ T cell response to cognate viral peptides
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7L9I
 
 | | Crystal structure of human ARH3-D314A bound to magnesium and ADP-ribose | | Descriptor: | ADP-ribose glycohydrolase ARH3, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Kurinov, I, Moss, J, Kim, I.K. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical analysis of human ADP-ribosyl-acceptor hydrolase 3 reveals the basis of metal selectivity and different roles for the two magnesium ions.
J.Biol.Chem., 296, 2021
|
|
1H8V
 
 | | The X-ray Crystal Structure of the Trichoderma reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Shaw, A, Ropp, T.H, Wu, S, Bott, R, Cameron, A.D, Stahlberg, J, Mitchinson, C, Jones, T.A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution
J.Mol.Biol., 308, 2001
|
|
7LT9
 
 | |
6CEW
 
 | | Segment AMMAAA from the low complexity domain of TDP-43, residues 321-326 | | Descriptor: | AMMAAA | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8BES
 
 | |
5C3S
 
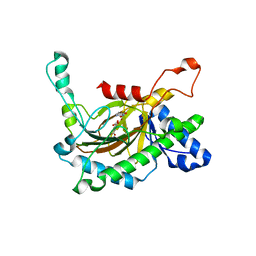 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-formyluracil (5fU) | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carbaldehyde, 2-OXOGLUTARIC ACID, ... | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
1H13
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
1PTM
 
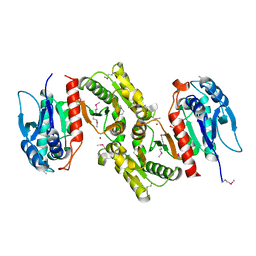 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, PHOSPHATE ION, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-23 | | Release date: | 2003-11-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
8IBN
 
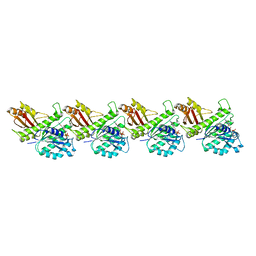 | | Cryo-EM structure of KpFtsZ single filament | | Descriptor: | Cell division protein FtsZ, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, POTASSIUM ION | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hibino, K, Konishi, T, Kato, Y, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
