5DR4
 
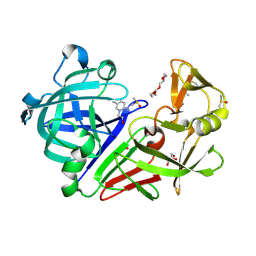 | | Endothiapepsin in complex with fragment 231 | | Descriptor: | 7-methyl-3,4-dihydro-2~{H}-pyrido[1,2-a]pyrimidin-3-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4OD0
 
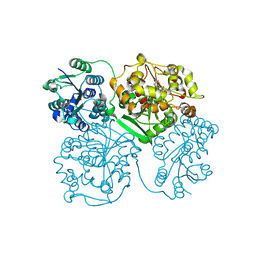 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
6R82
 
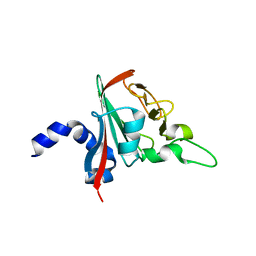 | |
7RZC
 
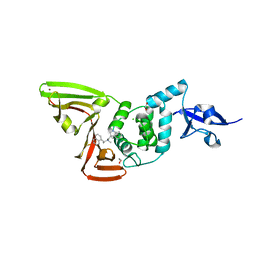 | | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor | | Descriptor: | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor
To be Published
|
|
6R5M
 
 | | Crystal structure of toxin MT9 from mamba venom | | Descriptor: | ACETYL GROUP, Dendroaspis polylepis MT9, GLYCEROL, ... | | Authors: | Stura, E.A, Tepshi, L, Ciolek, J, Triquigneaux, M, Zoukimian, C, De Waard, M, Beroud, R, Servent, D, Gilles, N, Legrand, P, Ciccone, L. | | Deposit date: | 2019-03-25 | | Release date: | 2020-02-12 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MT9, a natural peptide from black mamba venom antagonizes the muscarinic type 2 receptor and reverses the M2R-agonist-induced relaxation in rat and human arteries
Biomed Pharmacother, 150, 2022
|
|
7SDR
 
 | | Papain-Like Protease of SARS CoV-2 in Complex with Jun9-72-2 Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({methyl[(1R)-1-(naphthalen-1-yl)ethyl]amino}methyl)phenol, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in Complex with Jun9-72-2 Inhibitor
To be Published
|
|
7SA7
 
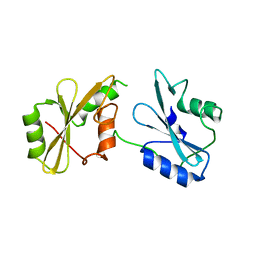 | |
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7SEV
 
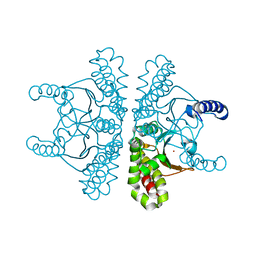 | | Crystal structure of E coli contaminant protein YadF co-purified with a plant protein | | Descriptor: | Carbonic anhydrase 2, POTASSIUM ION, ZINC ION | | Authors: | Chai, L, Zhu, P, Chai, J, Pang, C, Andi, B, McsWeeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2021-10-01 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AlphaFold Protein Structure Database for Sequence-Independent Molecular Replacement
Crystals, 11, 2021
|
|
7DFR
 
 | |
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4F2L
 
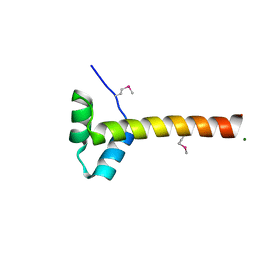 | | Structure of a regulatory domain of AMPK | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, MAGNESIUM ION | | Authors: | Xin, F.J, Zhang, Y.Y, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2012-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved regulatory elements in AMPK
Nature, 498, 2013
|
|
6D6V
 
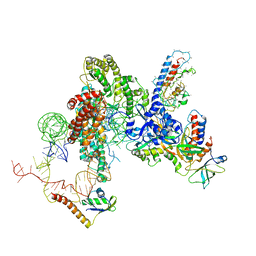 | | CryoEM structure of Tetrahymena telomerase with telomeric DNA at 4.8 Angstrom resolution | | Descriptor: | DNA (5'-D(P*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*G)-3'), RNA (159-MER), Telomerase associated protein p65, ... | | Authors: | Jiang, J, Wang, Y, Susac, L, Chan, H, Basu, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2018-04-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of Telomerase with Telomeric DNA.
Cell, 173, 2018
|
|
1HYK
 
 | | AGOUTI-RELATED PROTEIN (87-132) (AC-AGRP(87-132)) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Bolin, K.A, Anderson, D.J, Trulson, J.A, Thompson, D.A, Wilken, J, Kent, S.B.H, Millhauser, G.L. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a minimized human agouti related protein prepared by total chemical synthesis.
FEBS Lett., 451, 1999
|
|
1HZT
 
 | | CRYSTAL STRUCTURE OF METAL-FREE ISOPENTENYL DIPHOSPHATE:DIMETHYLALLYL DIPHOSPHATE ISOMERASE | | Descriptor: | ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE | | Authors: | Durbecq, V, Sainz, G, Oudjama, Y, Clantin, B, Bompard-Gilles, C, Tricot, C, Caillet, J, Stalon, V, Droogmans, L, Villeret, V. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase.
Embo J., 20, 2001
|
|
2XFL
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Sun, H, Bricogne, G, Liang, Z, Lescar, J. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
4TW9
 
 | | Difluoro-dioxolo-benzoimidazol-benzamides as potent inhibitors of CK1delta and epsilon with nanomolar inhibitory activity on cancer cell proliferation | | Descriptor: | CHLORIDE ION, Casein kinase I isoform delta, N-(2,2-difluoro-5H-[1,3]dioxolo[4,5-f]benzimidazol-6-yl)-2-{[2-(trifluoromethoxy)benzoyl]amino}-1,3-thiazole-4-carboxamide, ... | | Authors: | Richter, J, Bischof, J, Zaja, M, Kohlhof, H, Othersen, O, Vitt, D, Alscher, V, Pospiech, I, Garcia-Reyes, B, Berg, S, Leban, J, Knippschild, U. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Difluoro-dioxolo-benzoimidazol-benzamides As Potent Inhibitors of CK1 delta and epsilon with Nanomolar Inhibitory Activity on Cancer Cell Proliferation.
J.Med.Chem., 57, 2014
|
|
1I1G
 
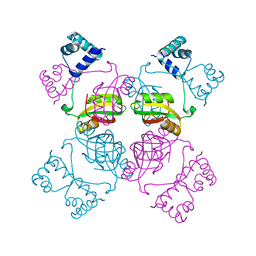 | | CRYSTAL STRUCTURE OF THE LRP-LIKE TRANSCRIPTIONAL REGULATOR FROM THE ARCHAEON PYROCOCCUS FURIOSUS | | Descriptor: | TRANSCRIPTIONAL REGULATOR LRPA | | Authors: | Leonard, P.M, Smits, S.H.J, Sedelnikova, S.E, Brinkman, A.B, de Vos, W.M, van der Oost, J, Rice, D.W, Rafferty, J.B. | | Deposit date: | 2001-02-01 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus.
EMBO J., 20, 2001
|
|
6DN3
 
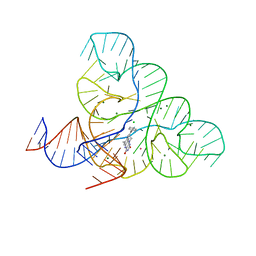 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1555 SPLIT RNA | | Descriptor: | 7,8-dimethyl-2,4-dioxo-10-(3-phenylpropyl)-1,2,3,4-tetrahydrobenzo[g]pteridin-10-ium, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DRZ
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N-[(2S)-1-hydroxybutan-2-yl]-1,6-dimethyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2XEM
 
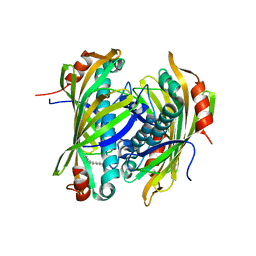 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | (3E,5E,7E,9E,11E,13E)-pentadeca-3,5,7,9,11,13-hexaen-2-one, DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Bricogne, G, Liang, Z.X, Lescar, J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
1I5L
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH SHORT POLY-U RNA | | Descriptor: | 5'-R(*UP*UP*U)-3', PUTATIVE SNRNP SM-LIKE PROTEIN AF-SM1, URIDINE | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-28 | | Release date: | 2001-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
4F7C
 
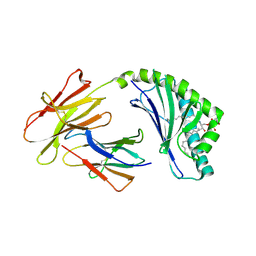 | | Crystal structure of bovine CD1d with bound C12-di-sulfatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CD1D antigen, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Plos One, 7, 2012
|
|
2XCD
 
 | | Structure of YncF,the genomic dUTPase from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Garcia, J, Burchell, L, Takezawa, M, Rzechorzek, N.J, Fogg, M, Wilson, K.S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
