5PFA
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 139) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PFQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 154) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PXB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 35) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3FCT
 
 | | MATURE METAL CHELATASE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Romesberg, F.E, Santarsiero, B.D, Barnes, D, Yin, J, Spiller, B, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic evidence for strain in biological catalysis.
Biochemistry, 37, 1998
|
|
5PXQ
 
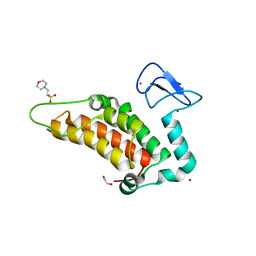 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 50) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3EXE
 
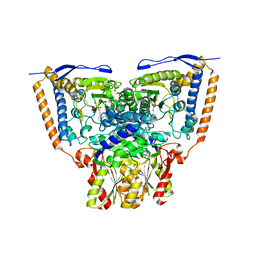 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
5PY7
 
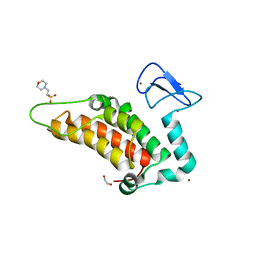 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 67) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4CKJ
 
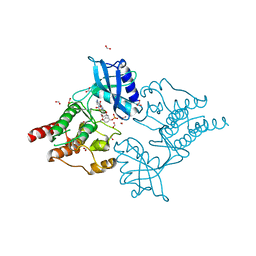 | | Crystal structure of RET tyrosine kinase domain bound to adenosine | | Descriptor: | ADENOSINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Goodman, K, Martinez-Torres, R.J, Borg, A, Murray-Rust, J, Mouilleron, S, Knowles, P, McDonald, N.Q. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Oncogenic RET kinase domain mutations perturb the autophosphorylation trajectory by enhancing substrate presentation in trans.
Mol. Cell, 53, 2014
|
|
3FYR
 
 | | Crystal structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis | | Descriptor: | Sporulation inhibitor sda | | Authors: | Jacques, D.A, Streamer, M, King, G.F, Guss, J.M, Trewhella, J, Langley, D.B. | | Deposit date: | 2009-01-23 | | Release date: | 2009-06-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5PG3
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 167) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3FZ0
 
 | | Inosine-Guanosine Nucleoside Hydrolase (IG-NH) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Nucleoside hydrolase, ... | | Authors: | Vandemeulebroucke, A, Minici, C, Bruno, I, Muzzolini, L, Tornaghi, P, Parkin, D.W, Schramm, V.L, Versees, W, Steyaert, J, Degano, M. | | Deposit date: | 2009-01-23 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the 6-oxopurine nucleosidase from Trypanosoma brucei brucei
Biochemistry, 49, 2010
|
|
3EZJ
 
 | | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody | | Descriptor: | CHLORIDE ION, General secretion pathway protein GspD, NANOBODY NBGSPD_7, ... | | Authors: | Korotkov, K.V, Pardon, E, Steyaert, J, Hol, W.G. | | Deposit date: | 2008-10-22 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody.
Structure, 17, 2009
|
|
5PGO
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 187) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PYJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 79) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4CQ4
 
 | | C-terminal fragment of Af1503-sol: transmembrane receptor Af1503 from Archaeoglobus fulgidus engineered for solubility | | Descriptor: | ENGINEERED VERSION OF TRANSMEMBRANE RECEPTOR AF1503 | | Authors: | Hartmann, M.D, Dunin-Horkawicz, S, Hulko, M, Martin, J, Coles, M, Lupas, A.N. | | Deposit date: | 2014-02-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Soluble Mutant of the Transmembrane Receptor Af1503 Features Strong Changes in Coiled-Coil Periodicity.
J.Struct.Biol., 186, 2014
|
|
3FE1
 
 | | Crystal structure of the human 70kDa heat shock protein 6 (Hsp70B') ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 6, ... | | Authors: | Wisniewska, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
5PZ4
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 100) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PZJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 115) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PH9
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09701a | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, N-OXALYLGLYCINE, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PHO
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 1) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3F0C
 
 | | Crystal structure of transcriptional regulator from Cytophaga hutchinsonii ATCC 33406 | | Descriptor: | SULFATE ION, Transcriptional regulator | | Authors: | Nocek, B, Maltseva, N, Tan, K, Abdullah, J, Eschenfeldt, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of transcriptional regulator from Cytophaga hutchinsonii ATCC 33406
To be Published
|
|
5PI4
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 17) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4CNV
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
5UPU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Mulligan, R, Gu, M, Sacchettini, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
5PIN
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 36) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
