6J0V
 
 | | Crystal Structure of Yeast Rtt107 | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J17
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ESX-3 secretion system protein EccC3, MAGNESIUM ION | | Authors: | Wang, S.H, Li, J, Rao, Z.H. | | Deposit date: | 2018-12-28 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
5KYM
 
 | | Crystal Structure of the 1-acyl-sn-glycerophosphate (LPA) acyltransferase, PlsC, from Thermotoga maritima | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, 1-acyl-sn-glycerol-3-phosphate acyltransferase, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Robertson, R.M, Yao, J, Gajewski, S, Kumar, G, Martin, E.W, Rock, C.O, White, S.W. | | Deposit date: | 2016-07-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2RJV
 
 | |
6J8I
 
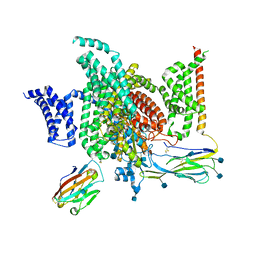 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (Y1755 up) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
6J9C
 
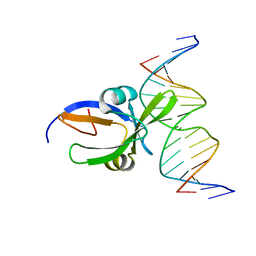 | | Crystal structure of Arabidopsis thaliana transcription factor LEC2-DNA complex | | Descriptor: | B3 domain-containing transcription factor LEC2, DNA (5'-D(*CP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
2RJX
 
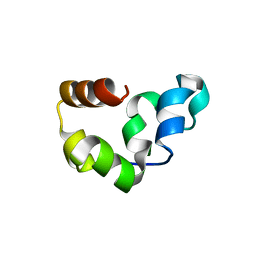 | |
2RH3
 
 | | Crystal structure of plasmid pTiC58 VirC2 | | Descriptor: | Protein virC2 | | Authors: | Lu, J, Glover, J.N.M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Agrobacterium tumefaciens VirC2 enhances T-DNA transfer and virulence through its C-terminal ribbon-helix-helix DNA-binding fold
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5KLP
 
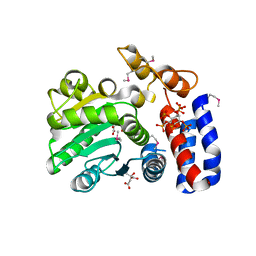 | | Crystal structure of HopZ1a in complex with IP6 | | Descriptor: | CITRIC ACID, INOSITOL HEXAKISPHOSPHATE, Orf34 | | Authors: | Zhang, Z.-M, Song, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-08-10 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7E4K
 
 | |
6JBG
 
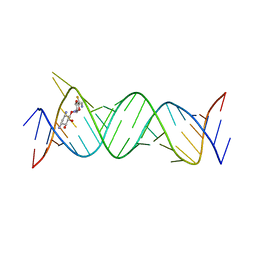 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (equatorial 4'-F) | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-glucopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3'), RNA (5'-R(P*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Auffinger, P, Hanessian, S, Kondo, J. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F)
To Be Published
|
|
5KOR
 
 | | Arabidopsis thaliana fucosyltransferase 1 (FUT1) in complex with GDP and a xylo-oligossacharide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rocha, J, de Sanctis, D, Breton, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation.
Plant Cell, 28, 2016
|
|
5KPQ
 
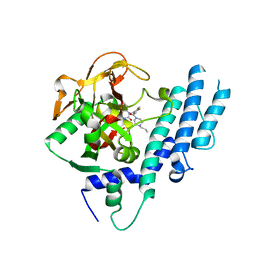 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-propyl-piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5KQ4
 
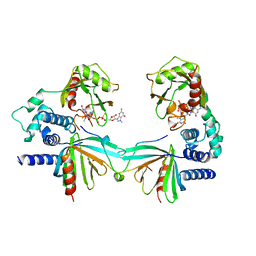 | | Crystal structure of S. pombe Dcp1/Dcp2 in complex with H. sapiens PNRC2 and synthetic cap analog | | Descriptor: | Proline-rich nuclear receptor coactivator 2, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] [[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-oxidanyl-phosphoryl] hydrogen phosphate, mRNA decapping complex subunit 2, ... | | Authors: | Mugridge, J.S, Ziemniak, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of mRNA-cap recognition by Dcp1-Dcp2.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5L8B
 
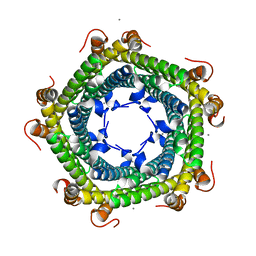 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5KQM
 
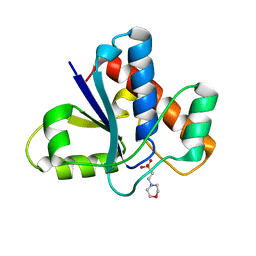 | | Co-crystal structure of LMW-PTP in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Wang, J, Zhang, Z.-Y, Yu, Z.-H. | | Deposit date: | 2016-07-06 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Inhibition of low molecular weight protein tyrosine phosphatase by an induced-fit mechanism.
J.Med.Chem., 2016
|
|
5LA8
 
 | |
2RMP
 
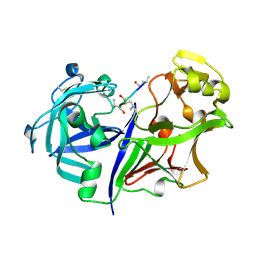 | | RMP-pepstatin A complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MUCOROPEPSIN, PEPSTATIN, ... | | Authors: | Yang, J, Quail, J.W. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Rhizomucor miehei aspartic proteinase complexed with the inhibitor pepstatin A at 2.7 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6J94
 
 | | Crystal structure of CYP97A3 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic | | Authors: | Niu, G, Guo, Q, Wang, J, Zhao, S. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5KRF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Dynamic WAY derivative, 1a | | Descriptor: | 4-[1-methyl-7-(trifluoromethyl)indazol-3-yl]benzene-1,3-diol, Estrogen receptor, KHKILHRLLQDSSS Peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6GRU
 
 | | Crystal structure of human NUDT5 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Fairhead, M, MacLean, E, Diaz Saez, L, Strain-Damerell, C, Elkins, J, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-12 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of human NUDT5
To Be Published
|
|
2RR3
 
 | | Solution structure of the complex between human VAP-A MSP domain and human OSBP FFAT motif | | Descriptor: | Oxysterol-binding protein 1, Vesicle-associated membrane protein-associated protein A | | Authors: | Furuita, K, Jee, J, Fukada, H, Mishima, M, Kojima, C. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies
J.Biol.Chem., 285, 2010
|
|
5KVB
 
 | |
5KW2
 
 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
2STB
 
 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
