8HVW
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
1C1C
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | Descriptor: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
6SD0
 
 | |
7ZWD
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4Z7U
 
 | | S13 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DQ-alpha chain, MHC class II HLA-DQ-beta-1, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H, Koning, F. | | Deposit date: | 2015-04-08 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
J Immunol., 194, 2015
|
|
7ZX7
 
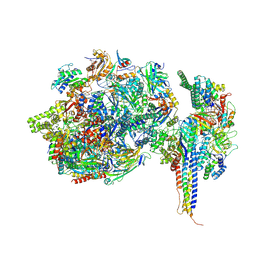 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1YCY
 
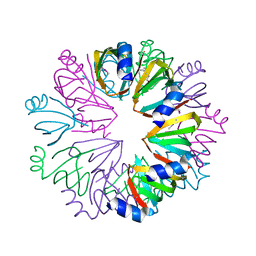 | | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein | | Authors: | Huang, L, Liu, Z.-J, Lee, D, Tempel, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus
To be published
|
|
7MY1
 
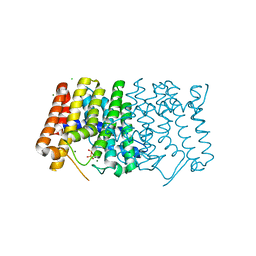 | | Sy-CrtE structure with IPP, N-term His-tag | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MXZ
 
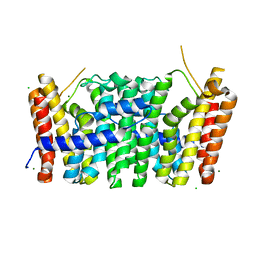 | | Sy-CrtE apo structure | | Descriptor: | CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7MY7
 
 | | Se-CrtE N-term His-tag structure | | Descriptor: | Farnesyl-diphosphate synthase | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
4R3M
 
 | | Crystal structure of Human Hsp90 with JR9 | | Descriptor: | Heat shock protein HSP 90-alpha, N~3~-benzyl-2-[(6-bromo-1,3-benzodioxol-5-yl)methyl]imidazo[1,2-a]pyrazine-3,8-diamine | | Authors: | Li, J, Yang, M, Ren, J, Xiong, B, He, J. | | Deposit date: | 2014-08-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multi-substituted 8-aminoimidazo[1,2-a]pyrazines by Groebke-Blackburn-Bienayme reaction and their Hsp90 inhibitory activity.
Org.Biomol.Chem., 13, 2015
|
|
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4Z7W
 
 | | T316 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQ8-glia-alpha1, MHC class II HLA-DQ-alpha chain, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H, Koning, F. | | Deposit date: | 2015-04-08 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
J Immunol., 194, 2015
|
|
7ZPJ
 
 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 isoform 1 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7MY6
 
 | | Se-CrtE C-term His-tag with IPP added | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
8HXS
 
 | | Small_spotted catshark CD8alpha | | Descriptor: | T-cell surface glycoprotein CD8 alpha chain | | Authors: | Wang, J, Zou, J. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The first crystal structure of CD8 alpha alpha from a cartilaginous fish.
Front Immunol, 14, 2023
|
|
4Z62
 
 | | The plant peptide hormone free receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine receptor 1 | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
1S1U
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
4Z5W
 
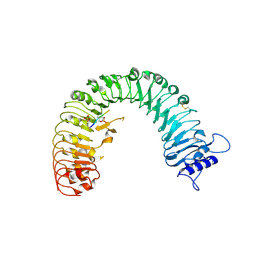 | | The plant peptide hormone receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine, ... | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
8A00
 
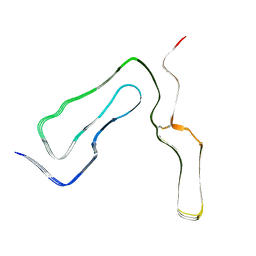 | | Infectious mouse-adapted ME7 scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for prion strain diversity.
Nat.Chem.Biol., 19, 2023
|
|
1S1W
 
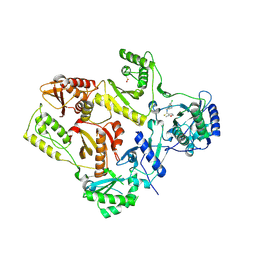 | | Crystal structure of V106A mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
7NM2
 
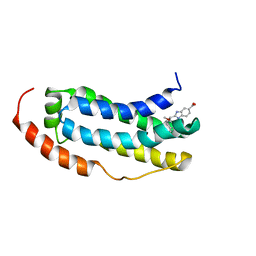 | | Solution structure of MLKL executioner domain in complex with a fragment | | Descriptor: | 2-[(~{S})-methoxy-(4-propan-2-ylphenyl)methyl]-3~{H}-benzimidazole-5-carboxylic acid, Mixed lineage kinase domain-like protein | | Authors: | Ruebbelke, M, Bauer, M, Hamilton, J, Binder, F, Nar, H, Zeeb, M. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Discovery and Structure-Based Optimization of Fragments Binding the Mixed Lineage Kinase Domain-like Protein Executioner Domain.
J.Med.Chem., 64, 2021
|
|
1C0U
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+50.0934 | | Descriptor: | (R)-(+) 5(9BH)-OXO-9B-PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-3-CARBOXYLIC ACID METHYL ESTER, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
7NM5
 
 | | Solution structure of MLKL executioner domain in complex with a fragment | | Descriptor: | 2-[(~{S})-methoxy-(4-phenylphenyl)methyl]-3~{H}-benzimidazole-5-carboxylic acid, Mixed lineage kinase domain-like protein | | Authors: | Ruebbelke, M, Bauer, M, Hamilton, J, Binder, F, Nar, H, Zeeb, M. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Discovery and Structure-Based Optimization of Fragments Binding the Mixed Lineage Kinase Domain-like Protein Executioner Domain.
J.Med.Chem., 64, 2021
|
|
