2AOD
 
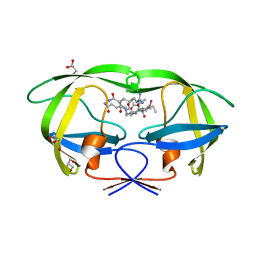 | | Crystal structure analysis of HIV-1 protease with a substrate analog P2-NC | | 分子名称: | DIMETHYL SULFOXIDE, GLYCEROL, HIV-1 PROTEASE, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
6FLR
 
 | |
2HMI
 
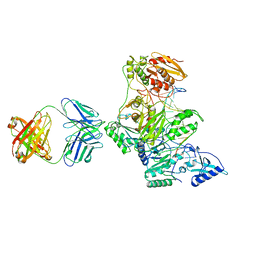 | | HIV-1 REVERSE TRANSCRIPTASE/FRAGMENT OF FAB 28/DNA COMPLEX | | 分子名称: | DNA (5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3'), FAB FRAGMENT OF MONOCLONAL ANTIBODY 28, ... | | 著者 | Ding, J, Arnold, E. | | 登録日 | 1998-04-10 | | 公開日 | 1998-10-14 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution.
J.Mol.Biol., 284, 1998
|
|
7EF1
 
 | |
7EF2
 
 | |
7EEZ
 
 | |
6FWB
 
 | | Crystal structure of Mat2A at 1.79 Angstron resolution | | 分子名称: | GLYCEROL, S-adenosylmethionine synthase isoform type-2, SODIUM ION, ... | | 著者 | Zhou, A, Wei, Z, Bai, J, Wang, H. | | 登録日 | 2018-03-06 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes.
Ebiomedicine, 39, 2019
|
|
7W1Y
 
 | | Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (49-MER), ... | | 著者 | Li, J, Dong, J, Dang, S, Zhai, Y. | | 登録日 | 2021-11-21 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | The human pre-replication complex is an open complex.
Cell, 186, 2023
|
|
7EF3
 
 | |
2B51
 
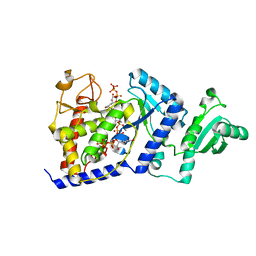 | | Structural Basis for UTP Specificity of RNA Editing TUTases from Trypanosoma Brucei | | 分子名称: | MANGANESE (II) ION, RNA editing complex protein MP57, URIDINE 5'-TRIPHOSPHATE | | 著者 | Deng, J, Ernst, N.L, Turley, S, Stuart, K.D, Hol, W.G. | | 登録日 | 2005-09-27 | | 公開日 | 2005-11-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
Embo J., 24, 2005
|
|
6GB9
 
 | |
5APG
 
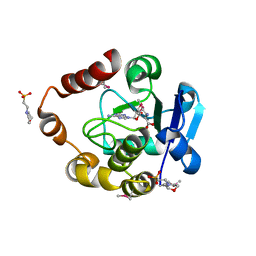 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | 著者 | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | 登録日 | 2015-09-15 | | 公開日 | 2016-04-27 | | 最終更新日 | 2016-06-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2013-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
6FR0
 
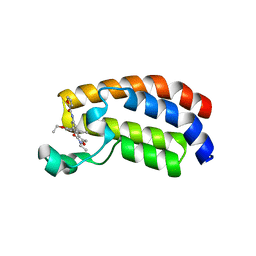 | | Crystal structure of CREBBP bromodomain complexd with PB08 | | 分子名称: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(2-ethyl-5-methyl-3-oxidanylidene-1,2-oxazol-4-yl)phenyl]furan-2-carboxamide | | 著者 | Zhu, J, Caflisch, A. | | 登録日 | 2018-02-15 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
1Y69
 
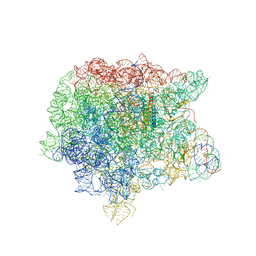 | | RRF domain I in complex with the 50S ribosomal subunit from Deinococcus radiodurans | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L16, 50S ribosomal protein L27, ... | | 著者 | Wilson, D.N, Schluenzen, F, Harms, J.M, Yoshida, T, Ohkubo, T, Albrecht, R, Buerger, J, Kobayashi, Y, Fucini, P. | | 登録日 | 2004-12-04 | | 公開日 | 2005-03-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | X-ray crystallography on ribosome recycling: mechanism of binding and action of RRF on the 50S ribosomal subunit
EMBO J., 24, 2005
|
|
7EF0
 
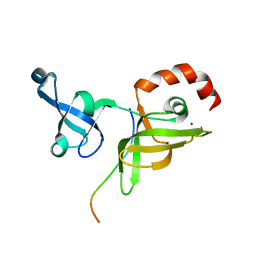 | |
6FRF
 
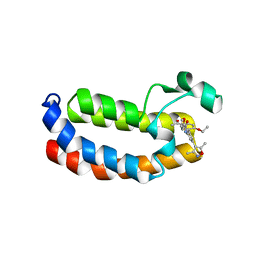 | | Crystal structure of CREBBP bromodomain complexd with PA10 | | 分子名称: | CREB-binding protein, ~{N}-[3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | 著者 | Zhu, J, Caflisch, A. | | 登録日 | 2018-02-15 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
3LKO
 
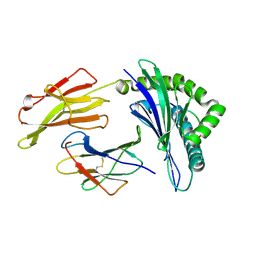 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1934 strain | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | 著者 | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | 登録日 | 2010-01-27 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8OO2
 
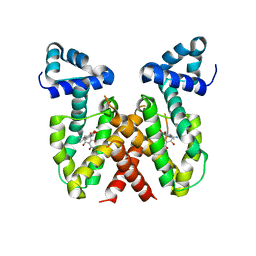 | | ChdA complex with amido-chelocardin | | 分子名称: | 2-carboxamido-2-deacetyl-chelocardin, MAGNESIUM ION, Putative transcriptional regulator | | 著者 | Koehnke, J, Sikandar, A. | | 登録日 | 2023-04-04 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Revision of the Absolute Configurations of Chelocardin and Amidochelocardin.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2I1Y
 
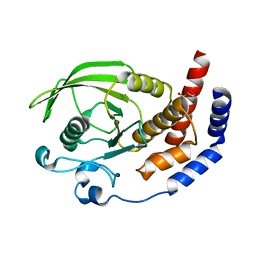 | | Crystal structure of the phosphatase domain of human PTP IA-2 | | 分子名称: | GLYCEROL, Receptor-type tyrosine-protein phosphatase | | 著者 | Faber-Barata, J, Patskovsky, Y, Alvarado, J, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Maletic, M, Rooney, I, Bain, K.T, Freeman, M, Russell, J.C, Thompson, D.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-08-15 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
7E24
 
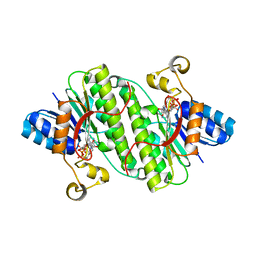 | |
7E28
 
 | |
4NAX
 
 | | Crystal structure of glutathione transferase PPUT_1760 from Pseudomonas putida, target EFI-507288, with one glutathione disulfide bound per one protein subunit | | 分子名称: | FORMIC ACID, GLYCEROL, Glutathione S-transferase, ... | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-10-22 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.301 Å) | | 主引用文献 | Crystal structure of glutathione transferase Pput_1760 from Pseudomonas putida, target EFI-507288
To be Published
|
|
5VEI
 
 | | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2 | | 分子名称: | Sorbin and SH3 domain-containing protein 2, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Huang, H, Gu, J, Liu, K, Sidhu, S.S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-04-04 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2
To be Published
|
|
5G4Y
 
 | | Structural basis for carboxylic acid recognition by a Cache chemosensory domain. | | 分子名称: | Methyl-accepting chemotaxis sensory transducer with Cache sensor, UNKNOWN LIGAND | | 著者 | Brewster, J, McKellar, J.L.O, Newman, J, Peat, T.S, Gerth, M.L. | | 登録日 | 2016-05-18 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae.
Sci Rep, 6, 2016
|
|
