7VLQ
 
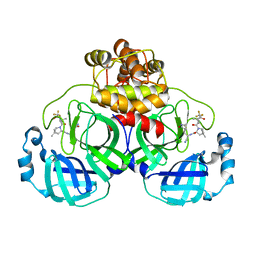 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.939106 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLO
 
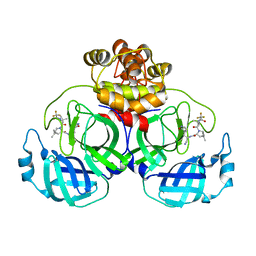 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.0227 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.50251937 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
6EJX
 
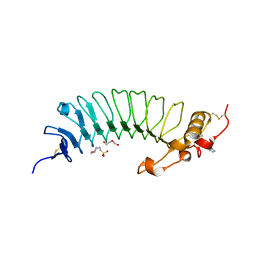 | |
4E08
 
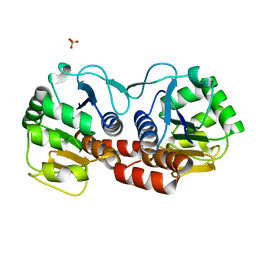 | |
4LRH
 
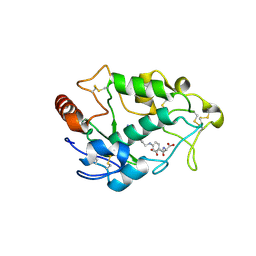 | | Crystal structure of human folate receptor alpha in complex with folic acid | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, Folate receptor alpha | | 著者 | Ke, J, Chen, C, Zhou, X.E, Yi, W, Brunzelle, J.S, Li, J, Young, E.-L, Xu, H.E, Melcher, K. | | 登録日 | 2013-07-19 | | 公開日 | 2013-07-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for molecular recognition of folic acid by folate receptors.
Nature, 500, 2013
|
|
1XY1
 
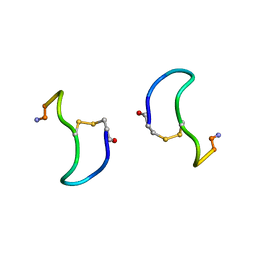 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | 分子名称: | BETA-MERCAPTOPROPIONATE-OXYTOCIN | | 著者 | Husain, J, Blundell, T.L, Wood, S.P, Tickle, I.J, Cooper, S, Pitts, J.E. | | 登録日 | 1987-06-05 | | 公開日 | 1988-04-16 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
3HMW
 
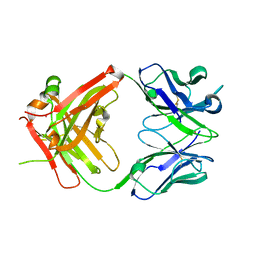 | | Crystal structure of ustekinumab FAB | | 分子名称: | CADMIUM ION, USTEKINUMAB FAB HEAVY CHAIN, USTEKINUMAB FAB LIGHT CHAIN | | 著者 | Luo, J. | | 登録日 | 2009-05-29 | | 公開日 | 2010-06-09 | | 最終更新日 | 2015-07-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab.
J.Mol.Biol., 402, 2010
|
|
1MAX
 
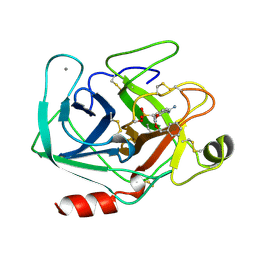 | | BETA-TRYPSIN PHOSPHONATE INHIBITED | | 分子名称: | BETA-TRYPSIN, CALCIUM ION, [N-(BENZYLOXYCARBONYL)AMINO](4-AMIDINOPHENYL)METHANE-PHOSPHONATE | | 著者 | Bertrand, J, Oleksyszyn, J, Kam, C, Boduszek, B, Presnell, S, Plaskon, R, Suddath, F, Powers, J, Williams, L. | | 登録日 | 1996-02-06 | | 公開日 | 1996-10-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Inhibition of trypsin and thrombin by amino(4-amidinophenyl)methanephosphonate diphenyl ester derivatives: X-ray structures and molecular models.
Biochemistry, 35, 1996
|
|
4M1Q
 
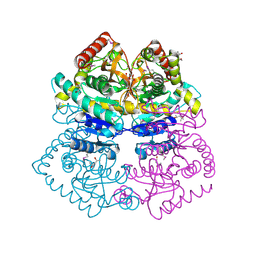 | | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-lactate dehydrogenase, PHOSPHATE ION | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-08-03 | | 公開日 | 2013-08-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814.
To be Published
|
|
4DGK
 
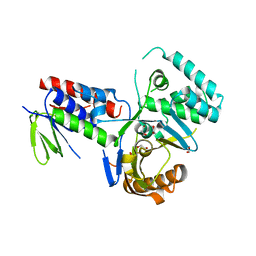 | | Crystal structure of Phytoene desaturase CRTI from Pantoea ananatis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Phytoene dehydrogenase | | 著者 | Schaub, P, Yu, Q, Gemmecker, S, Poussin-Courmontagne, P, Mailliot, J, McEwen, A.G, Ghisla, S, Beyer, P, Cavarelli, J. | | 登録日 | 2012-01-26 | | 公開日 | 2012-10-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | On the structure and function of the phytoene desaturase CRTI from Pantoea ananatis, a membrane-peripheral and FAD-dependent oxidase/isomerase.
Plos One, 7, 2012
|
|
5B8D
 
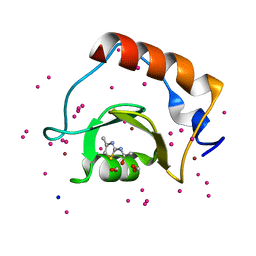 | | Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | 分子名称: | FORMIC ACID, Histone deacetylase 6, SODIUM ION, ... | | 著者 | Harding, R.J, Tempel, W, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Ravichandran, M, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | 登録日 | 2016-06-14 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
6G87
 
 | | Flavonoid-responsive Regulator FrrA | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TetR/AcrR family transcriptional regulator | | 著者 | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | 登録日 | 2018-04-07 | | 公開日 | 2019-04-24 | | 最終更新日 | 2021-08-18 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
4LYB
 
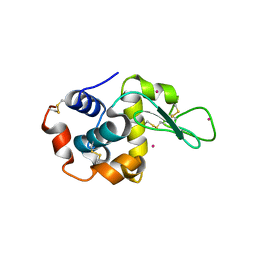 | | CdS within a lysoyzme single crystal | | 分子名称: | CADMIUM ION, Lysozyme C | | 著者 | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | 登録日 | 2013-07-30 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
4HCZ
 
 | | PHF1 Tudor in complex with H3K36me3 | | 分子名称: | H3L-like histone, PHD finger protein 1 | | 著者 | Musselman, C.A, Roy, S, Nunez, J, Kutateladze, T.G. | | 登録日 | 2012-10-01 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1XYN
 
 | |
4MXD
 
 | | 1.45 angstronm crystal structure of E.coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) | | 分子名称: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | 登録日 | 2013-09-26 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
7YC7
 
 | | Dark, fully reduced structure of the MmCPDII-DNA complex as produced at SwissFEL | | 分子名称: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2022-07-01 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
6QKB
 
 | | Crystal structure of the beta-hydroxyaspartate aldolase of Paracoccus denitrificans | | 分子名称: | D-3-hydroxyaspartate aldolase, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Zarzycki, J, Schada von Borzyskowski, L, Gilardet, A, Erb, T.J. | | 登録日 | 2019-01-28 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Marine Proteobacteria metabolize glycolate via the beta-hydroxyaspartate cycle.
Nature, 575, 2019
|
|
7YD7
 
 | | TR-SFX MmCPDII-DNA complex: 1 ns snapshot. Includes 1 ns, dark, and extrapolated structure factors | | 分子名称: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2022-07-04 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
4N18
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342 | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CITRIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase family protein | | 著者 | Bacal, P, Shabalin, I.G, Cooper, D.R, Majorek, K.A, Osinski, T, Hillerich, B.S, Hammonds, J, Nawar, A, Stead, M, Chowdhury, S, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-10-03 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342
To be Published
|
|
7YCM
 
 | | TR-SFX MmCPDII-DNA complex: 100 ps snapshot. Includes 100ps, dark, and extrapolated structure factors | | 分子名称: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2022-07-01 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCR
 
 | | TR-SFX MmCPDII-DNA complex: 450 ps snapshot. Includes 450ps, dark, and extrapolated structure factors | | 分子名称: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2022-07-01 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
5AP8
 
 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | 分子名称: | TSR3 | | 著者 | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | 登録日 | 2015-09-14 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.246 Å) | | 主引用文献 | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
7YCP
 
 | | TR-SFX MmCPDII-DNA complex: 250 ps snapshot. Includes 250 ps, dark, and extrapolated structure factors | | 分子名称: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2022-07-01 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
