5CVN
 
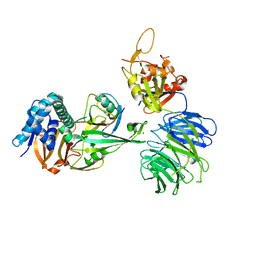 | | WDR48 (2-580):USP46~ubiquitin ternary complex | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 46, WD repeat-containing protein 48, ... | | Authors: | Harris, S.F, Yin, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
3C0F
 
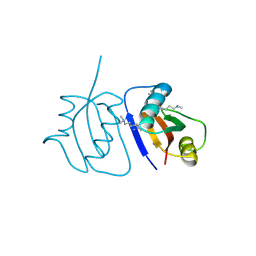 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
1GKQ
 
 | |
8ODZ
 
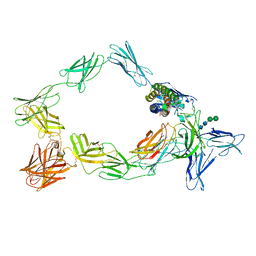 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, Interleukin-12 receptor subunit beta-2,Calmodulin-1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5CWU
 
 | | Crystal structure of Chaetomium thermophilum Nup188 TAIL domain | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
3HME
 
 | | Crystal structure of human bromodomain containing 9 isoform 1 (BRD9) | | Descriptor: | Bromodomain-containing protein 9 | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Picaud, S, Roos, A, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
3HRO
 
 | | Crystal structure of a C-terminal coiled coil domain of Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2, polycystic kidney disease 2) | | Descriptor: | Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2), also called Polycystin-2 or polycystic kidney disease 2(PKD2) | | Authors: | Yu, Y, Ulbrich, M.H, Li, M.-H, Buraei, Z, Chen, X.-Z, Ong, A.C.M, Tong, L, Isacoff, E.Y, Yang, J. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular basis of the assembly of the TRPP2/PKD1 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5IQO
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitutions Q134E and S138E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_T4R-T6R-D13N | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Giese, C, Eras, J, Kern, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8OE4
 
 | | Cryo-EM structure of a pre-dimerized human IL-23 complete extracellular signaling complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Bloch, Y, Felix, J, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5OAR
 
 | | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Skerlova, J, Rezacova, P, Brynda, J, Pachl, P, Otwinowski, Z, Vanek, O. | | Deposit date: | 2017-06-23 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae sheds light onto its substrate specificity, high stability, and regulation by propeptide.
FEBS J., 285, 2018
|
|
3C3N
 
 | | Crystal structure of dihydroorotate dehydrogenase from Trypanosoma cruzi strain Y | | Descriptor: | Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Pinheiro, M.P, Iulek, J, Nonato, M.C. | | Deposit date: | 2008-01-28 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase from Y strain
Biochem.Biophys.Res.Commun., 369, 2008
|
|
8OE0
 
 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 2). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7XYF
 
 | |
7XYG
 
 | |
7XXE
 
 | |
3C5M
 
 | | Crystal structure of oligogalacturonate lyase (VPA0088) from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR199 | | Descriptor: | MANGANESE (II) ION, Oligogalacturonate lyase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Janjua, H, Mao, L, Xiao, R, Owens, L.A, Wang, D, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of oligogalacturonate lyase (VPA0088) from Vibrio parahaemolyticus.
To be Published
|
|
7K7V
 
 | | The X-ray crystal structure of SSR4, an S. pombe chromatin remodelling protein: iodide derivative | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Ssr4, a Schizosaccharomyces pombe chromatin-remodelling protein.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1R65
 
 | | Crystal structure of ferrous soaked Ribonucleotide Reductase R2 subunit (wildtype) at pH 5 from E. coli | | Descriptor: | FE (II) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
3HPH
 
 | | Closed tetramer of Visna virus integrase (residues 1-219) in complex with LEDGF IBD | | Descriptor: | GLYCEROL, Integrase, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Wang, J, Cherepanov, P. | | Deposit date: | 2009-06-04 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for functional tetramerization of lentiviral integrase
Plos Pathog., 5, 2009
|
|
3BWC
 
 | | Crystal structure of spermidine synthase from Trypanosoma cruzi in complex with SAM at 2.3 A resolution | | Descriptor: | BETA-MERCAPTOETHANOL, S-ADENOSYLMETHIONINE, Spermidine synthase | | Authors: | Bosch, J, Arakaki, T.L, Le Trong, I, Merritt, E.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2008-01-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of spermidine synthase from Trypanosoma cruzi.
To be Published
|
|
5OGN
 
 | | Metalacarborane inhibitors of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, ZINC ION, cobaltcarborane | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K, Kugler, M, Gruner, B, Sicha, V. | | Deposit date: | 2017-07-13 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Metallacarborane Sulfamides: Unconventional, Specific, and Highly Selective Inhibitors of Carbonic Anhydrase IX.
J.Med.Chem., 2019
|
|
8JYR
 
 | |
3C8T
 
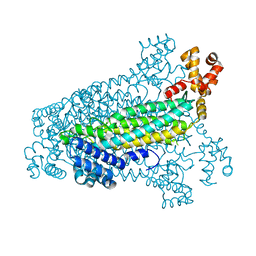 | | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1 | | Descriptor: | Fumarate lyase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-13 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1.
To be Published
|
|
8JYQ
 
 | |
