3ONL
 
 | | yeast Ent3_ENTH-Vti1p_Habc complex structure | | Descriptor: | Epsin-3, t-SNARE VTI1 | | Authors: | Wang, J, Fang, P, Niu, L, Teng, M. | | Deposit date: | 2010-08-29 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Epsin N-terminal homology domains bind on opposite sides of two SNAREs
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7RHD
 
 | | darkmRuby M94T/F96Y mutant at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby M94T/F96Y mutant | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
8BE9
 
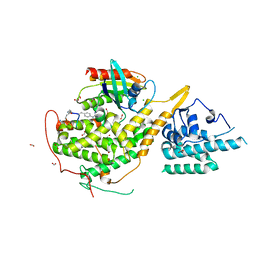 | | Crystal structure of SOS1-HRas-peptidomimetic5 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GTPase HRas, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7RDV
 
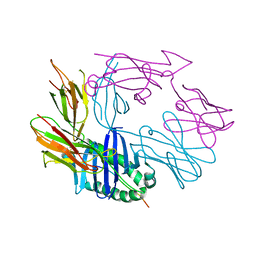 | |
2O9K
 
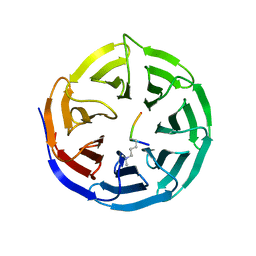 | | WDR5 in Complex with Dimethylated H3K4 Peptide | | Descriptor: | H3 HISTONE, WD repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Molecular Recognition and Presentation of Histone H3 by Wdr5.
Embo J., 25, 2006
|
|
8BEA
 
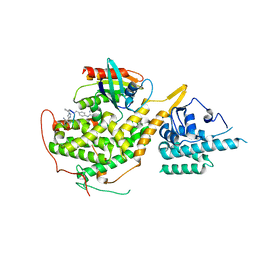 | |
8BE6
 
 | | Crystal structure of SOS1-HRas-peptidomimetic2 | | Descriptor: | GTPase HRas, SOS1-HRas-peptidomimetic2, Son of sevenless homolog 1 | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.89880252 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6WRX
 
 | | Crystal structure of computationally designed protein 2DS25.1 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RHC
 
 | | A new fluorescent protein darkmRuby at pH 9.0 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
6QWK
 
 | | The Transcriptional Regulator PrfA-L140F mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6WRV
 
 | | Crystal structure of computationally designed protein 3DS18 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 3DS18, ... | | Authors: | Abraham, J, Baker, D, Sahtoe, D.D, Coscia, A, Clark, L, Olal, D. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8BNU
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) tetrameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
6WJZ
 
 | |
6WRW
 
 | | Crystal structure of computationally designed protein 2DS25.5 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 2DS25.5, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8BRJ
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) trimeric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
6QMT
 
 | | Complement factor D in complex with the inhibitor 2-(2-(3'-(aminomethyl)-[1,1'-biphenyl]-3-carboxamido)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
6QN5
 
 | |
6QYJ
 
 | |
6W0A
 
 | | Open-gate KcsA soaked in 1 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.237 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0I
 
 | | Closed-gate KcsA soaked in 10mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6QNL
 
 | |
3ONK
 
 | | yeast Ent3_ENTH domain | | Descriptor: | Epsin-3 | | Authors: | Wang, J, Fang, P, Niu, L, Teng, M. | | Deposit date: | 2010-08-29 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Epsin N-terminal homology domains bind on opposite sides of two SNAREs
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8BVW
 
 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
6W9Q
 
 | | Peptide-bound SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | 3C-like proteinase peptide, Non-structural protein 9 fusion, PHOSPHATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
6W66
 
 | | The structure of the F64A, S172A mutant Keap1-BTB domain in complex with SKP1-FBXL17 | | Descriptor: | F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, S-phase kinase-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
