4N5B
 
 | | Crystal structure of the Nipah virus phosphoprotein tetramerization domain | | Descriptor: | IMIDAZOLE, Phosphoprotein | | Authors: | Bruhn, J.F, Barnett, K, Bibby, J, Thomas, J, Keegan, R, Rigden, D, Bornholdt, Z.A, Saphire, E.O. | | Deposit date: | 2013-10-09 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the nipah virus phosphoprotein tetramerization domain.
J.Virol., 88, 2014
|
|
4RLO
 
 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
7LW2
 
 | |
4EAM
 
 | | 1.70A resolution structure of apo beta-glycosidase (W33G) from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Deckert, K, Brunner, L.C, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designing allosteric control into enzymes by chemical rescue of structure.
J.Am.Chem.Soc., 134, 2012
|
|
7LYX
 
 | | Crystal structure of human CYP8B1 in complex with (S)-tioconazole | | Descriptor: | (S)-Tioconazole, 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase, GLYCEROL, ... | | Authors: | Liu, J, Scott, E.E. | | Deposit date: | 2021-03-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and characterization of human cytochrome P450 8B1 supports future drug design for nonalcoholic fatty liver disease and diabetes.
J.Biol.Chem., 298, 2022
|
|
7M0G
 
 | |
7LER
 
 | | Netrin-1 filament assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Netrin-1, ... | | Authors: | McDougall, M, Gupta, M, Stetefeld, J. | | Deposit date: | 2021-01-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.99 Å) | | Cite: | The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation.
Nat Commun, 14, 2023
|
|
1UCN
 
 | | X-ray structure of human nucleoside diphosphate kinase A complexed with ADP at 2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Chen, Y, Gallois-Montbrun, S, Schneider, B, Veron, M, Morera, S, Deville-Bonne, D, Janin, J. | | Deposit date: | 2003-04-16 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide Binding to Nucleoside Diphosphate Kinases: X-ray Structure of Human NDPK-A in Complex with ADP and Comparison to Protein Kinases
J.Mol.Biol., 332, 2003
|
|
4EPH
 
 | | CRYSTAL STRUCTURE OF RAT CARNITINE PALMITOYLTRANSFERASE 2 IN COMPLEX with CoA-site inhibitor | | Descriptor: | 2-chloro-4-[({1-[(5-chloro-2-methoxyphenyl)sulfonyl]-4-methyl-2,3-dihydro-1H-indol-6-yl}carbonyl)amino]benzoic acid, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2012-04-17 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
7MDN
 
 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
4EFI
 
 | | Crystal Structure of 3-oxoacyl-(Acyl-carrier protein) Synthase from Burkholderia Xenovorans LB400 | | Descriptor: | 3-oxoacyl-(Acyl-carrier protein) synthase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Craig, T.K, Abendroth, J, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
4EQ4
 
 | | Crystal structure of seleno-methionine derivatized GH3.12 | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Zubieta, C, Nanao, M, Jez, J, Westfall, C, Kapp, U. | | Deposit date: | 2012-04-18 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
1UHY
 
 | | Crystal structure of d(GCGATAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*TP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
1UI1
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, Uracil-DNA Glycosylase | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
1UHX
 
 | | Crystal structure of d(GCGAGAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
1UI0
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, URACIL, ... | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
4EP9
 
 | | CRYSTAL STRUCTURE OF RAT CARNITINE PALMITOYLTRANSFERASE 2 IN COMPLEX WITH CoA-site inhibitor | | Descriptor: | 4-[({1-[(5-chloro-2-methoxyphenyl)sulfonyl]-4-methyl-2,3-dihydro-1H-indol-6-yl}carbonyl)amino]benzoic acid, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2012-04-17 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
1UIU
 
 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel unliganded form) | | Descriptor: | Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
7MFR
 
 | | Crystal Structure of a Fab fragment bound to peptide GGM | | Descriptor: | Antibody fragment - Heavy Chain of fab, Antibody fragment - Light Chain of fab, GLY-GLY-MET, ... | | Authors: | Sudhamsu, J. | | Deposit date: | 2021-04-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Antibody toolkit reveals N-terminally ubiquitinated substrates of UBE2W.
Nat Commun, 12, 2021
|
|
1UIV
 
 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel liganded form) | | Descriptor: | NICKEL (II) ION, Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
7MBR
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (apo state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBS
 
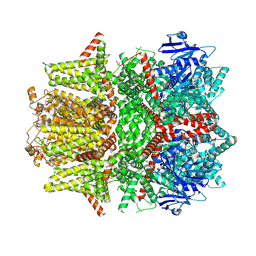 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (open state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MWM
 
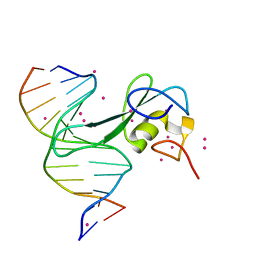 | | Crystal structure of MBD2 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*A)-R(P*(5MC))-D(P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of MBD2 with DNA
To Be Published
|
|
7MBT
 
 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (low calcium occupancy in the transmembrane domain) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MWK
 
 | | Crystal structure of MBD2 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(MC)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Crystal structure of MBD2 with DNA
To Be Published
|
|
