5H99
 
 | |
6GA6
 
 | | Bacteriorhodopsin, 10 ps state, real-space refined against 10% extrapolated map | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
5H9I
 
 | | Crystal structure of Geobacter metallireducens SMUG1 with xanthine | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Geobacter metallireducens SMUG1, ... | | Authors: | Xie, W, Cao, W, Zhang, Z, Shen, J. | | Deposit date: | 2015-12-28 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural Basis of Substrate Specificity in Geobacter metallireducens SMUG1
Acs Chem.Biol., 11, 2016
|
|
6GAG
 
 | | BACTERIORHODOPSIN, 630 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
5GMB
 
 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
4J2F
 
 | | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866 | | Descriptor: | GLYCEROL, Glutathione s-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-02-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866
TO BE PUBLISHED
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
6C69
 
 | | Structure of glycolipid aGSA[12,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | Descriptor: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | Authors: | Zhang, X, Ma, J, Wang, Y, Wang, J. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
5GRB
 
 | | Crystal structure of 2C helicase from enterovirus 71 (EV71) bound with ATPgammaS | | Descriptor: | EV71 2C ATPase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ZINC ION | | Authors: | Guan, H.X, Tian, J, Qin, B, Wojdyla, J, Wang, M.T, Cui, S. | | Deposit date: | 2016-08-09 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of 2C helicase from enterovirus 71
SCI ADV, 3, 2017
|
|
5M7V
 
 | | Translation initiation factor 4E in complex with (RP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D1) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(sulfanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Translation initiation factor 4E in complex with (RP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D1)
To Be Published
|
|
5HJN
 
 | |
5HLG
 
 | | Structure of reduced AbfR bound to DNA | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*TP*CP*AP*AP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*TP*GP*AP*GP*T)-3'), MarR family transcriptional regulator | | Authors: | Liu, G, Liu, X, Gan, J, Yang, C.G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Redox-Sensing Mechanism of MarR-Type Regulator AbfR.
J. Am. Chem. Soc., 139, 2017
|
|
1YLJ
 
 | | Atomic resolution structure of CTX-M-9 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-9a | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1YM9
 
 | | Crystal structure of the CDC25B phosphatase catalytic domain with the active site cysteine in the sulfinic form | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2 | | Authors: | Buhrman, G.K, Parker, B, Sohn, J, Rudolph, J, Mattos, C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Mechanism of Oxidative Regulation of the Phosphatase Cdc25B via an Intramolecular Disulfide Bond
Biochemistry, 44, 2005
|
|
6CXF
 
 | |
5LS2
 
 | | Receptor mediated chitin perception in legumes is functionally seperable from Nod factor perception | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, SULFATE ION | | Authors: | Bozsoki, Z, Cheng, J, Feng, F, Gysel, K, Andersen, K.R, Oldroyd, G, Blaise, M, Radutoiu, S, Stougaard, J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
1YML
 
 | | Crystal Structure of the CDC25B phosphatase catalytic domain with the active site cysteine in the sulfenic form | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2 | | Authors: | Buhrman, G.K, Parker, B, Sohn, J, Rudolph, J, Mattos, C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Mechanism of Oxidative Regulation of the Phosphatase Cdc25B via an Intramolecular Disulfide Bond
Biochemistry, 44, 2005
|
|
5H85
 
 | |
5LDW
 
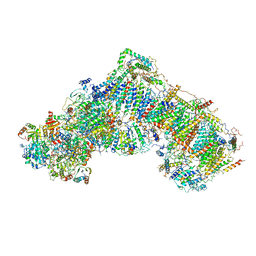 | | Structure of mammalian respiratory Complex I, class1 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
1YLP
 
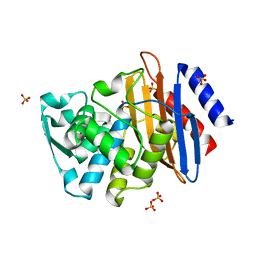 | | Atomic resolution structure of CTX-M-27 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-27 | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
5JV0
 
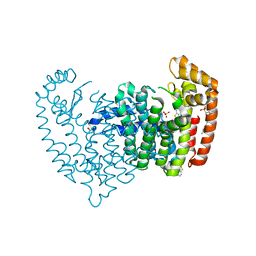 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-08-038 | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION, [(1R)-2-(3-fluorophenyl)-1-{[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}ethyl]phosphonic acid | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
5LI2
 
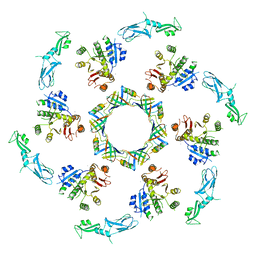 | | bacteriophage phi812K1-420 tail sheath and tail tube protein in native tail | | Descriptor: | Phage-like element PBSX protein XkdM, tail sheath protein | | Authors: | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | Deposit date: | 2016-07-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7X45
 
 | | Grass carp interferon gamma related | | Descriptor: | Interferon gamma | | Authors: | Wang, J, Zou, J, Zhu, X. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-14 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel Dimeric Architecture of an IFN-gamma-Related Cytokine Provides Insights into Subfunctionalization of Type II IFNs in Teleost Fish.
J Immunol., 209, 2022
|
|
