7F8T
 
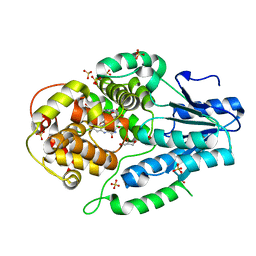 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
6HMG
 
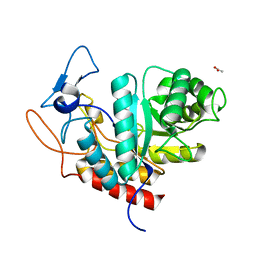 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
4BED
 
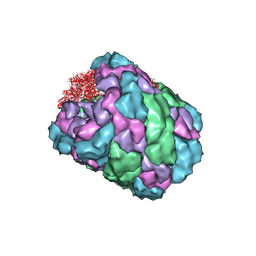 | |
2MVF
 
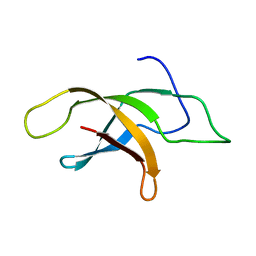 | | Structural insight into an essential assembly factor network on the pre-ribosome | | Descriptor: | Uncharacterized protein | | Authors: | Lee, W, Bassler, J, Paternoga, H, Holdermann, I, Thomas, M, Granneman, S, Barrio-Garcia, C, Nyarko, A, Stier, G, Clark, S.A, Schraivogel, D, Kallas, M, Beckmann, R, Tollervey, D, Barbar, E, Sinning, I, Hurt, E. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
1DSB
 
 | |
5LC0
 
 | | Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor | | Descriptor: | N-((S)-3-(4-(aminomethyl)phenyl)-1-(((R)-4-guanidino-1-(5-hydroxy-1,3,2-dioxaborinan-2-yl)butyl)amino)-1-oxopropan-2-yl)benzamide, NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Lei, J, Hansen, G, Zhang, L.L, Hilgenfeld, R. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor.
Science, 353, 2016
|
|
2MX8
 
 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 7.2 | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
2V05
 
 | |
5LV5
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1458 | | Descriptor: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]-[(3~{S})-3-azanyl-4-oxidanyl-4-oxidanylidene-butyl]azanium, Protein arginine N-methyltransferase 6 | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
2UZX
 
 | | Structure of the human receptor tyrosine kinase Met in complex with the Listeria monocytogenes invasion protein InlB: Crystal form I | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR, INTERNALIN B | | Authors: | Niemann, H.H, Jager, V, Butler, P.J.G, Van Den Heuvel, J, Schmidt, S, Ferraris, D, Gherardi, E, Heinz, D.W. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Human Receptor Tyrosine Kinase met in Complex with the Listeria Invasion Protein Inlb
Cell(Cambridge,Mass.), 130, 2007
|
|
5LUF
 
 | | Cryo-EM of bovine respirasome | | Descriptor: | COPPER (II) ION, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Sousa, J.S, Mills, D.J, Vonck, J, Kuehlbrandt, W. | | Deposit date: | 2016-09-08 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Functional asymmetry and electron flow in the bovine respirasome.
Elife, 5, 2016
|
|
2VEW
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | 3-fluoro-N-[(1S)-1-[4-[(2-fluorophenyl)methyl]imidazol-2-yl]-2-[4-[(5S)-1,1,3-trioxo-1,2-thiazolidin-5-yl]phenyl]ethyl]benzenesulfonamide, MAGNESIUM ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VAX
 
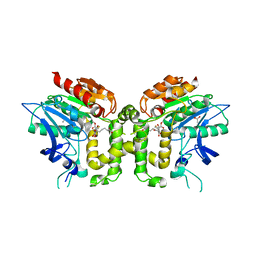 | | Crystal structure of deacetylcephalosporin C acetyltransferase (Cephalosporin C-soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
5TS8
 
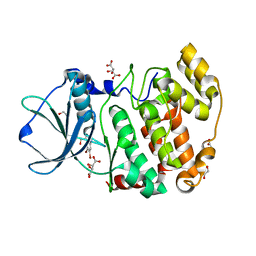 | | Z. MAYS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIBROMOBENZOTRIAZOLE, ACETATE ION, ... | | Authors: | Winiewska, M, Kucinska, K, Czapinska, H, Piasecka, A, Bochtler, M, Poznanski, J. | | Deposit date: | 2016-10-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | STRUCTURAL AND THERMODYNAMIC ANALYSIS OF 5,6-DIBROMOBENZOTRIAZOLE BINDING TO CASEIN KINASE 2 ALPHA
To Be Published
|
|
5LZQ
 
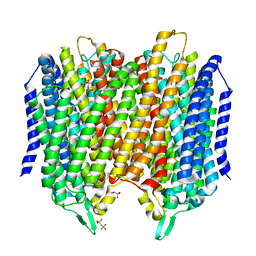 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sodium ion | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Wilkinson, C, Kellosalo, J, Kajander, T, Goldman, A. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Membrane pyrophosphatases from Thermotoga maritima and Vigna radiata suggest a conserved coupling mechanism.
Nat Commun, 7, 2016
|
|
2MOP
 
 | | Structure of Bitistatin A | | Descriptor: | Disintegrin bitistatin | | Authors: | Carbajo, R.J, Calvete, J, Sanz, L, Perez, A. | | Deposit date: | 2014-04-29 | | Release date: | 2014-11-12 | | Last modified: | 2015-02-25 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bitistatin - a missing piece in the evolutionary pathway of snake venom disintegrins.
Febs J., 282, 2015
|
|
2V4R
 
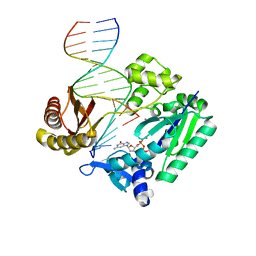 | | Non-productive complex of the Y-family DNA polymerase Dpo4 with dGTP skipping the M1dG adduct to pair with the next template cytosine | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3', 5'-D(*TP*CP*AP*CP*M1GP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Stafford, J.B, Szekely, J, Rizzo, C.J, Egli, M, Guengerich, F.P, Marnett, L.J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of Sulfolobus Solfataricus Y-Family DNA Polymerase Dpo4-Catalyzed Bypass of the Malondialdehyde-Deoxyguanosine Adduct.
Biochemistry, 48, 2009
|
|
4E55
 
 | |
5M61
 
 | |
2W46
 
 | | CBM35 from Cellvibrio japonicus Abf62 | | Descriptor: | CALCIUM ION, ESTERASE D, SODIUM ION | | Authors: | Montainer, C, Lammerts van Bueren, A, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-01-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1FN9
 
 | | CRYSTAL STRUCTURE OF THE REOVIRUS OUTER CAPSID PROTEIN SIGMA 3 | | Descriptor: | OUTER-CAPSID PROTEIN SIGMA 3, ZINC ION | | Authors: | Olland, A.M, Jane-Valbuena, J, Schiff, L.A, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2000-08-21 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 A resolution.
EMBO J., 20, 2001
|
|
6HXZ
 
 | |
2W05
 
 | | Structure of CDK2 in complex with an imidazolyl pyrimidine, compound 5b | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(2-METHOXYETHYL)-4-({4-[2-METHYL-1-(1-METHYLETHYL)-1H-IMIDAZOL-5-YL]PYRIMIDIN-2-YL}AMINO)BENZENESULFONAMIDE | | Authors: | Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Breed, J, Byth, K.F, Culshaw, J.D, Finlay, M.R, Fisher, E, Green, C.P, Heaton, D.W, Nash, I.A, Newcombe, N.J, Oakes, S.E, Pauptit, R.A, Roberts, A, Stanway, J.J, Thomas, A.P, Tucker, J.A, Weir, H.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Imidazoles: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5MDJ
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in a its as-isolated high-pressurized form | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2W0D
 
 | | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? A Case Study of Metalloproteinases. | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Isaksson, J, Nystrom, S, Derbyshire, D.J, Wallberg, H, Agback, T, Kovacs, H, Bertini, I, Felli, I.C. | | Deposit date: | 2008-08-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? a Case Study of Metalloproteinases.
J.Med.Chem., 52, 2009
|
|
