4D6K
 
 | | Structure of DNTTIP1 dimerisation domain. | | Descriptor: | DEOXYNUCLEOTIDYLTRANSFERASE TERMINAL-INTERACTING PROTEIN 1 | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Cell Cycle Associated Hdac1/2 Complex Reveals the Structural Basis for Complex Assembly and Nucleosome Targeting.
Nucleic Acids Res., 43, 2015
|
|
6K5J
 
 | | Structure of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GH3 beta-N-acetylglucosaminidase, GLYCEROL | | Authors: | Itoh, T, Araki, T, Nishiyama, T, Hibi, T, Kimoto, H. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural and functional characterization of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7.
J.Biochem., 166, 2019
|
|
2FUZ
 
 | | UGL hexagonal crystal structure without glycine and DTT molecules | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2FV0
 
 | | UGL_D88N/dGlcA-Glc-Rha-Glc | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-beta-D-glucopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-beta-D-glucopyranose, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2FV1
 
 | | UGL_D88N/dGlcA-GlcNAc | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2GH4
 
 | | YteR/D143N/dGalA-Rha | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-03-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate
Biochem.Biophys.Res.Commun., 347, 2006
|
|
6K0M
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11 | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, GLYCEROL, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0U
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0Q
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0N
 
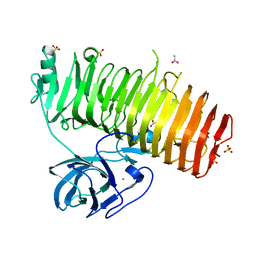 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0P
 
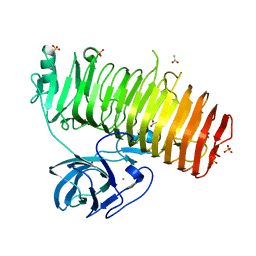 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0V
 
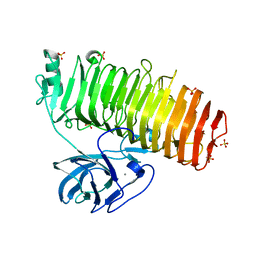 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0S
 
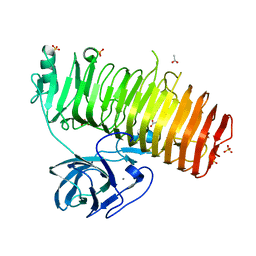 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
5ZEQ
 
 | | Carboxypeptidase B in complex with DD28 | | Descriptor: | (2~{S})-2-[(6-azanyl-5-chloranyl-pyridin-3-yl)methyl]-3-selanyl-propanoic acid, CACODYLATE ION, Carboxypeptidase B, ... | | Authors: | Itoh, T, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the selective inhibition of activated thrombin-activatable fibrinolysis inhibitor (TAFIa) by a selenium-containing inhibitor with chloro-aminopyridine as a basic group
Bioorg. Med. Chem. Lett., 28, 2018
|
|
2VST
 
 | | hPPARgamma Ligand binding domain in complex with 13-(S)-HODE | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV3
 
 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV0
 
 | | hPPARgamma Ligand binding domain in complex with DHA | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV2
 
 | | hPPARgamma Ligand binding domain in complex with 5-HEPA | | Descriptor: | (5R,6E,8Z,11Z,14Z,17Z)-5-hydroxyicosa-6,8,11,14,17-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV4
 
 | | hPPARgamma Ligand binding domain in complex with 6-oxoOTE | | Descriptor: | (8E,10S,12Z)-10-hydroxy-6-oxooctadeca-8,12-dienoic acid, (8R,9Z,12Z)-8-hydroxy-6-oxooctadeca-9,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2ZBL
 
 | | Functional annotation of Salmonella enterica yihS-encoded protein | | Descriptor: | Putative isomerase, beta-D-mannopyranose | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
3A09
 
 | |
3AX3
 
 | |
3AX2
 
 | | Crystal structure of rat TOM20-ALDH presequence complex: a disulfide-tethered complex with a non-optimized, long linker | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
3AX5
 
 | | Crystal structure of rat TOM20-ALDH presequence complex: A complex (form1) between Tom20 and a disulfide-bridged presequence peptide containing D-Cys and L-Cys at the i and i+3 positions. | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
5XUQ
 
 | | Crystal structure of VDR-LBD complexed with an antagonist, 2-methylidene-19,26,27-trinor-22-(S)-butyl-1-hydroxy-25-oxo-25-(1H-pyrrol-2-yl)- vitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(1~{H}-pyrrol-2-yl)octan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-24 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent Vitamin D Receptor Antagonist
To Be Published
|
|
