2IPC
 
 | | Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer | | Descriptor: | Preprotein translocase SecA subunit | | Authors: | Vassylyev, D.G, Mori, H, Vassylyeva, M.N, Tsukazaki, T, Kimura, Y, Tahirov, T.H, Ito, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Translocation ATPase SecA from Thermus thermophilus Reveals a Parallel, Head-to-Head Dimer.
J.Mol.Biol., 364, 2006
|
|
1IRE
 
 | | Crystal Structure of Co-type nitrile hydratase from Pseudonocardia thermophila | | Descriptor: | COBALT (II) ION, Nitrile Hydratase | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Wakagi, T. | | Deposit date: | 2001-10-01 | | Release date: | 2002-10-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cobalt-containing nitrile hydratase.
Biochem.Biophys.Res.Commun., 288, 2001
|
|
1J2T
 
 | | Creatininase Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, ZINC ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1J2U
 
 | | Creatininase Zn | | Descriptor: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1KOL
 
 | | Crystal structure of formaldehyde dehydrogenase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ZINC ION, ... | | Authors: | Tanaka, N, Kusakabe, Y, Ito, K, Yoshimoto, T, Nakamura, K.T. | | Deposit date: | 2001-12-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Formaldehyde Dehydrogenase from Pseudomonas putida: the Structural Origin of the Tightly Bound Cofactor in Nicotinoprotein Dehydrogenases
J.mol.biol., 324, 2002
|
|
5FG3
 
 | | Crystal structure of GDP-bound aIF5B from Aeropyrum pernix | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Probable translation initiation factor IF-2 | | Authors: | Murakami, R, Miyoshi, T, Uchiumi, T, Ito, K. | | Deposit date: | 2015-12-20 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of translation initiation factor 5B from the crenarchaeon Aeropyrum pernix.
Proteins, 84, 2016
|
|
3VMF
 
 | | Archaeal protein | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Saito, K, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2011-12-12 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex
Nucleic Acids Res., 40, 2012
|
|
6J3L
 
 | | Solution structure of the N-terminal extended protuberant domain of eukaryotic ribosomal stalk protein P0 | | Descriptor: | 60S acidic ribosomal protein P0 | | Authors: | Choi, K.H.A, Lee, K.M, Yang, L, Wing-Heng Yu, C, Banfield, D.K, Ito, K, Uchiumi, T, Wong, K.B. | | Deposit date: | 2019-01-04 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mutagenesis Studies Evince the Role of the Extended Protuberant Domain of Ribosomal Protein uL10 in Protein Translation.
Biochemistry, 58, 2019
|
|
2Z3W
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A | | Descriptor: | Dipeptidyl aminopeptidase IV, GLYCEROL, SULFATE ION | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-07 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
2Z3Z
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A complexd with an inhibitor | | Descriptor: | Dipeptidyl aminopeptidase IV, SULFATE ION, [(2R)-1-(L-ALANYL-L-ISOLEUCYL)PYRROLIDIN-2-YL]BORONIC ACID | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-09 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
6JI2
 
 | | Crystal structure of archaeal ribosomal protein aP1, aPelota, and GTP-bound aEF1A complex | | Descriptor: | Archaeal ribosomal stalk protein aP1, Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Maruyama, K, Imai, H, Kawamura, M, Ishino, S, Ishino, Y, Ito, K, Uchiumi, T. | | Deposit date: | 2019-02-20 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Switch of the interactions between the ribosomal stalk and EF1A in the GTP- and GDP-bound conformations.
Sci Rep, 9, 2019
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
2ZTU
 
 | | T190A mutant of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
2ZTM
 
 | | T190S mutant of D-3-hydroxybutyrate dehydrogenase | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
3A6J
 
 | | E122Q mutant creatininase complexed with creatine | | Descriptor: | Creatinine amidohydrolase, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6D
 
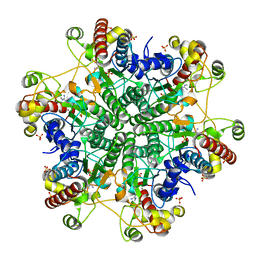 | | Creatininase complexed with 1-methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
2ZXG
 
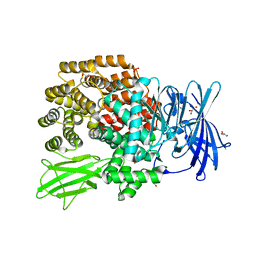 | | Aminopeptidase N complexed with the aminophosphinic inhibitor of PL250, a transition state analogue | | Descriptor: | Aminopeptidase N, GLYCEROL, N-{(2S)-3-[(1R)-1-aminoethyl](hydroxy)phosphoryl-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli complexed with the transition-state analogue aminophosphinic inhibitor PL250
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZTL
 
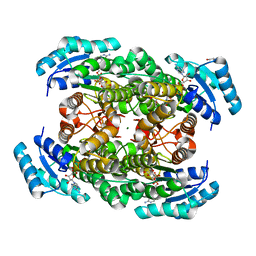 | | Closed conformation of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ and L-3-hydroxybutyrate | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, GLYCEROL, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
2ZTV
 
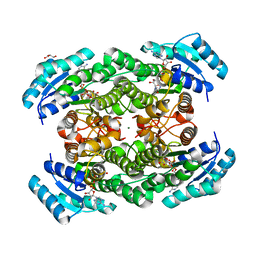 | | The binary complex of D-3-hydroxybutyrate dehydrogenase with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
3A6E
 
 | | W174F mutant creatininase, type I | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6L
 
 | | E122Q mutant creatininase, Zn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6H
 
 | | W154A mutant creatininase | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3AGK
 
 | | Crystal structure of archaeal translation termination factor, aRF1 | | Descriptor: | Peptide chain release factor subunit 1 | | Authors: | Kobayashi, K, Kikuno, I, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Omnipotent role of archaeal elongation factor 1 alpha (EF1{alpha}) in translational elongation and termination, and quality control of protein synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
