4MU3
 
 | | The form A structure of an E21Q catalytic mutant of A. thaliana IGPD2 in complex with Mn2+ and a mixture of its substrate, 2R3S-IGP, and an inhibitor, 2S3S-IGP, to 1.12 A resolution | | Descriptor: | (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, (2S,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, 1,2-ETHANEDIOL, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4MU1
 
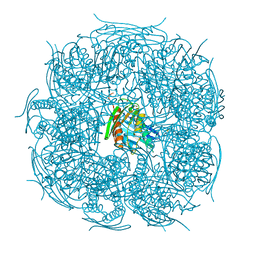 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+, imidazole, and sulfate at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
3CJ3
 
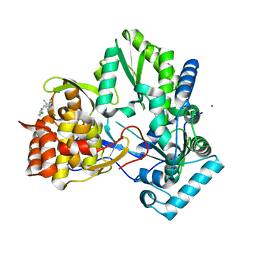 | |
4MU4
 
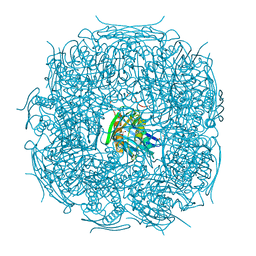 | | The form B structure of an E21Q catalytic mutant of A. thaliana IGPD2 in complex with Mn2+ and its substrate, 2R3S-IGP, to 1.41 A resolution | | Descriptor: | (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4MU0
 
 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and 1,2,4-triazole at 1.3 A resolution | | Descriptor: | 1,2,4-TRIAZOLE, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
1AUI
 
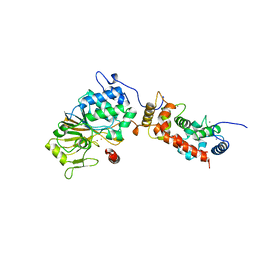 | | HUMAN CALCINEURIN HETERODIMER | | Descriptor: | CALCIUM ION, FE (III) ION, SERINE/THREONINE PHOSPHATASE 2B, ... | | Authors: | Kissinger, C.R, Parge, H.E, Knighton, D.R, Pelletier, L.A, Lewis, C.T, Tempczyk, A, Villafranca, J.E. | | Deposit date: | 1997-08-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex.
Nature, 378, 1995
|
|
4J03
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with fulvestrant | | Descriptor: | (7beta,9beta,13alpha,17beta)-7-{9-[(R)-(4,4,5,5,5-pentafluoropentyl)sulfinyl]nonyl}estra-1(10),2,4-triene-3,17-diol, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Morisseau, C, Pakhomova, S, Hwang, S.H, Newcomer, M.E, Hammock, B.D. | | Deposit date: | 2013-01-30 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Inhibition of soluble epoxide hydrolase by fulvestrant and sulfoxides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6HUD
 
 | |
6GHT
 
 | | HtxB D206A protein variant from Pseudomonas stutzeri in complex with hypophosphite to 1.12 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2018-05-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
6EMN
 
 | | HtxB from Pseudomonas stutzeri in complex with phosphite to 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable phosphite transport system-binding protein HtxB, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2017-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
6GHQ
 
 | | HtxB D206N protein variant from Pseudomonas stutzeri in a partially open conformation to 1.53 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
1U7T
 
 | | Crystal Structure of ABAD/HSD10 with a Bound Inhibitor | | Descriptor: | 1-AZEPAN-1-YL-2-PHENYL-2-(4-THIOXO-1,4-DIHYDRO-PYRAZOLO[3,4-D]PYRIMIDIN-5-YL)ETHANONE ADDUCT, 3-hydroxyacyl-CoA dehydrogenase type II, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kissinger, C.R, Rejto, P.A, Pelletier, L.A, Showalter, R.E, Villafranca, J.E. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human ABAD/HSD10 with a bound inhibitor: implications for design of Alzheimer's disease therapeutics
J.Mol.Biol., 342, 2004
|
|
6YS9
 
 | |
6YSG
 
 | |
6YT0
 
 | |
6YTJ
 
 | |
6YTN
 
 | |
7NR2
 
 | | The structure of the SBP TarP_Sse in complex with coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NRR
 
 | | The structure of the SBP TarP_Csal in complex with caffeate | | Descriptor: | CAFFEIC ACID, MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-04 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NQG
 
 | | The structure of the SBP TarP_Rhp in complex with 4-hydroxyphenylacetate | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENYLACETATE, PHOSPHATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NRA
 
 | | The structure of the SBP TarP_Sse in complex with cinnamate | | Descriptor: | HYDROCINNAMIC ACID, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NSW
 
 | | The structure of the SBP TarP_Csal in complex with coumarate | | Descriptor: | 1,2-ETHANEDIOL, 4'-HYDROXYCINNAMIC ACID, MAGNESIUM ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTD
 
 | | The structure of the SBP TarP_Csal in complex with ferulate | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
5VJB
 
 | |
5VJ9
 
 | |
