6TR1
 
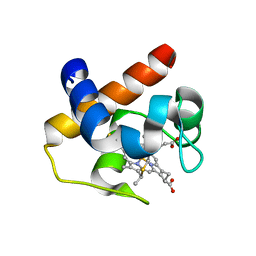 | | Native cytochrome c6 from Thermosynechococcus elongatus in space group H3 | | Descriptor: | Cytochrome c6, HEME C, SODIUM ION | | Authors: | Falke, S, Feiler, C.G, Sarrou, I. | | Deposit date: | 2019-12-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of native cytochrome c6from Thermosynechococcus elongatus in two different space groups and implications for its oligomerization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4URE
 
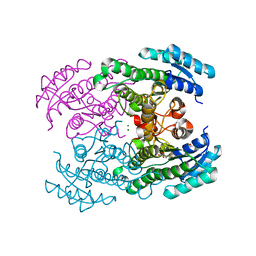 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, CYCLOHEXANOL DEHYDROGENASE, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
6TU4
 
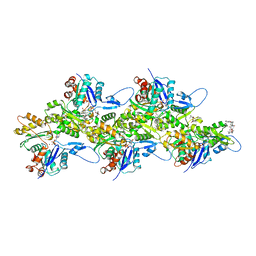 | | Structure of Plasmodium Actin1 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Vahokoski, J, Calder, L.J, Lopez, A.J, Rosenthal, P.B, Kursula, I. | | Deposit date: | 2020-01-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution structures of malaria parasite actomyosin and actin filaments.
Plos Pathog., 18, 2022
|
|
6TJH
 
 | | Crystal structure of the computationally designed Cake9 protein | | Descriptor: | Cake9, GLYCEROL, SULFATE ION | | Authors: | Mylemans, B, Laier, I, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
4V18
 
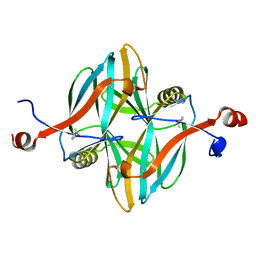 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6TG8
 
 | | Crystal structure of the Kelch domain in complex with 11 amino acid peptide (model of the ETGE loop) | | Descriptor: | Kelch-like ECH-associated protein 1, SODIUM ION, VAL-ILE-ASN-PRO-GLU-THR-GLY-GLU-GLN-ILE-GLN | | Authors: | Kekez, I, Matic, S, Tomic, S, Matkovic-Calogovic, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Binding of dipeptidyl peptidase III to the oxidative stress cell sensor Kelch-like ECH-associated protein 1 is a two-step process.
J.Biomol.Struct.Dyn., 39, 2021
|
|
4V1L
 
 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6TGF
 
 | | Pantoea stewartii WceF is a glycan biofilm modifying enzyme with a bacteriophage tailspike-like parallel beta-helix fold | | Descriptor: | 1,2-ETHANEDIOL, Exopolysaccharide biosynthesis protein, TETRAETHYLENE GLYCOL | | Authors: | Irmscher, T, Roske, Y, Gayk, I, Heinemann, U, Barbirz, S. | | Deposit date: | 2019-11-15 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Pantoea stewartii WceF is a glycan biofilm-modifying enzyme with a bacteriophage tailspike-like fold.
J.Biol.Chem., 296, 2021
|
|
6TI4
 
 | | SHMT from Streptococcus thermophilus Tyr55Ser variant in complex with PLP/D-Serine/Lys230 gem diamine complex | | Descriptor: | (2~{R})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2019-11-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of Y55S Serine Hydroxymethyltransferase variant from Streptococcus thermophilus in complex with gem-diamine intermediate of D-serine
To Be Published
|
|
6TJC
 
 | | Crystal structure of the computationally designed Cake3 protein | | Descriptor: | Cake3, GLYCEROL, PHOSPHATE ION | | Authors: | Laier, I, Mylemans, B, Voet, A.R.D, Noguchi, H. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJG
 
 | | Crystal structure of the computationally designed Cake8 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake8 | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TA5
 
 | | OprM-MexA complex from the MexAB-OprM Pseudomonas aeruginosa whole assembly reconstituted in nanodiscs | | Descriptor: | Efflux pump membrane transporter, MexA family multidrug efflux RND transporter periplasmic adaptor subunit, Outer membrane protein OprM | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
6TGH
 
 | | SHMT from Streptococcus thermophilus Tyr55Thr variant in complex with D-Serine both as external aldimine and as non-covalent complex | | Descriptor: | D-SERINE, L-Serine, N-[[3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]-4-pyridinyl]methylene], ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2019-11-15 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights into nucleophile substrate specificity in variants of N-Serine hydroxymethyltransferase from Streptococcus thermophilus
To Be Published
|
|
6THB
 
 | | Receptor binding domain of the Cedar Virus attachment glycoprotein (G) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein | | Authors: | Pryce, R, Rissanen, I, Harlos, K, Bowden, T. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus.
Life Sci Alliance, 3, 2020
|
|
4UZN
 
 | | The native structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B) | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
4V1K
 
 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | 2-HYDROXY BUTANE-1,4-DIOL, CALCIUM ION, CARBOHYDRATE BINDING MODULE, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6TOV
 
 | | Crystal Structure of Teicoplanin Aglycone | | Descriptor: | DIMETHYL SULFOXIDE, Teicoplanin Aglycone | | Authors: | Belviso, B.D, Carrozzini, B, Caliandro, R, Altomare, C.D, Bolognino, I, Cellamare, S. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (0.767 Å) | | Cite: | Enantiomeric Separation and Molecular Modelling of Bioactive 4-Aryl-3,4-dihydropyrimidin-2(1H)-one Ester Derivatives on Teicoplanin-Based Chiral Stationary Phase
Separations, 2022
|
|
6TSY
 
 | | Cytochrome c6 from Thermosynechococcus elongatus in a monoclinic space group | | Descriptor: | Cytochrome c6, HEME C, SULFATE ION | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2019-12-21 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of native cytochrome c6from Thermosynechococcus elongatus in two different space groups and implications for its oligomerization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TU7
 
 | | Structure of PfMyoA decorated Plasmodium Act1 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Vahokoski, J, Calder, L.J, Lopez, A.J, Rosenthal, P.B, Kursula, I. | | Deposit date: | 2020-01-03 | | Release date: | 2021-01-13 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution structures of malaria parasite actomyosin and actin filaments.
Plos Pathog., 18, 2022
|
|
6TI3
 
 | | Apo-SHMT from Streptococcus thermophilus Tyr55Ser variant in complex with D-Threonine | | Descriptor: | D-THREONINE, GLYCEROL, SODIUM ION, ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2019-11-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into nucleophile substrate specificity in variants of N-Serine hydroxymethyltransferase from Streptococcus thermophilus
To Be Published
|
|
4UZ8
 
 | | The SeMet structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B), SULFATE ION | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2015-05-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
6TRL
 
 | | Human Eg5 motor domain mutant Y82F | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Kinesin-like protein KIF11, ... | | Authors: | Garcia-Saez, I, Skoufias, D.A. | | Deposit date: | 2019-12-19 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human Eg5 motor domain mutant Y82F
To Be Published
|
|
6TLE
 
 | | Human Eg5 motor domain mutant E344K | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Garcia-Saez, I, Skoufias, D.A. | | Deposit date: | 2019-12-02 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human Eg5 motor domain mutant E344K
To Be Published
|
|
6TPB
 
 | | NMR structure of the apo-form of Pseudomonas fluorescens CopC | | Descriptor: | Putative copper resistance protein | | Authors: | Persson, K.C, Mayzel, M, Karlsson, B.G, Peciulyte, A, Olsson, L, Wittung Stafshede, P, Salomon Johansen, K, Horvath, I. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Pseudomonas fluorescens CopC
To Be Published
|
|
6TX3
 
 | | HPF1 bound to catalytic fragment of PARP2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 2,Poly [ADP-ribose] polymerase 2 | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
