3RXU
 
 | | Crystal structure of Trypsin complexed with benzamide (F05 and A06, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXO
 
 | | Crystal structure of Trypsin complexed with (3-pyrrol-1-ylphenyl)methanamine | | Descriptor: | 1-[3-(1H-pyrrol-1-yl)phenyl]methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3ZXP
 
 | |
3S0E
 
 | | Apis mellifera OBP14 in complex with the odorant eugenol (2-methoxy-4(2-propenyl)-phenol) | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3ZZI
 
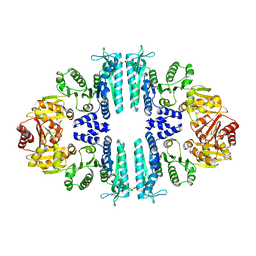 | | Crystal structure of a tetrameric acetylglutamate kinase from Saccharomyces cerevisiae | | Descriptor: | ACETYLGLUTAMATE KINASE | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
3RXL
 
 | | Crystal structure of Trypsin complexed with (2,5-dimethyl-3-furyl)methanamine | | Descriptor: | 1-(2,5-dimethylfuran-3-yl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXI
 
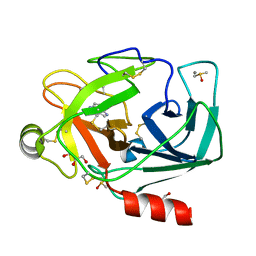 | | Crystal structure of Trypsin complexed with 2-(1H-indol-3-yl)ethanamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3SOP
 
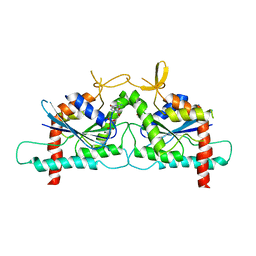 | | Crystal Structure Of Human Septin 3 GTPase Domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Neuronal-specific septin-3 | | Authors: | Marques, I.A, Macedo, J.N.A, Pereira, H.M, Valadares, N.F, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2011-06-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.885 Å) | | Cite: | The structure and properties of septin 3: a possible missing link in septin filament formation.
Biochem.J., 450, 2013
|
|
3ZFL
 
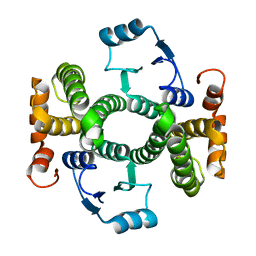 | | Crystal structure of the V58A mutant of human class alpha glutathione transferase in the apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Parbhoo, N, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Gildenhuys, S, Dirr, H.W. | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the V58A Mutant of Human Class Alpha Glutathione Transferase in the Apo Form
To be Published
|
|
3SZ1
 
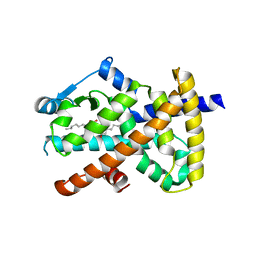 | | Human PPAR gamma ligand binding domain in complex with luteolin and myristic acid | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, MYRISTIC ACID, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Puhl, A.C, Bernardes, A, Polikarpov, I. | | Deposit date: | 2011-07-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mode of peroxisome proliferator-activated receptor gamma activation by luteolin.
Mol.Pharmacol., 81, 2012
|
|
3ZG2
 
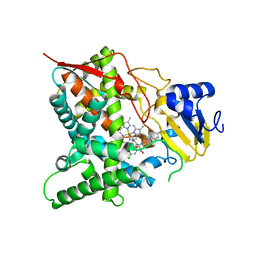 | | Sterol 14 alpha-demethylase (CYP51) from Trypanosoma cruzi in complex with the pyridine inhibitor (S)-2-(4-chlorophenyl)-2-(pyridin-3-yl)-1- (4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)ethanone (EPL-BS1246,UDO) | | Descriptor: | (S)-2-(4-chlorophenyl)-2-pyridin-3-yl-1-[4-[4-(trifluoromethyl)phenyl]piperazin-1-yl]ethanone, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Keenan, M, Chatelain, E, Lepesheva, G.I. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexes of Trypanosoma Cruzi Sterol 14Alpha-Demethylase (Cyp51) with Two Pyridine-Based Drug Candidates for Chagas Disease: Structural Basis for Pathogen-Selectivity
J.Biol.Chem., 288, 2013
|
|
3T3O
 
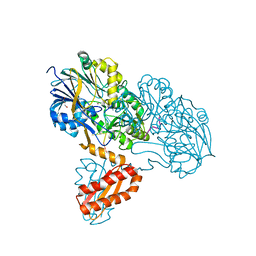 | | Molecular basis for the recognition and cleavage of RNA (CUGG) by the bifunctional 5'-3' exo/endoribonuclease RNase J | | Descriptor: | GLYCEROL, Metal dependent hydrolase, O2'methyl-RNA, ... | | Authors: | Dorleans, A, Li de la Sierra-Gallay, I, Piton, J, Zig, L, Gilet, L, Putzer, H, Condon, C. | | Deposit date: | 2011-07-25 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Recognition and Cleavage of RNA by the Bifunctional 5'-3' Exo/Endoribonuclease RNase J.
Structure, 19, 2011
|
|
4GHJ
 
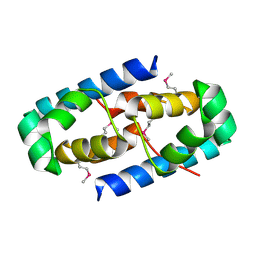 | | 1.75 Angstrom Crystal Structure of Transcriptional Regulator ftom Vibrio vulnificus. | | Descriptor: | Probable transcriptional regulator | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Crystal Structure of Transcriptional Regulator ftom Vibrio vulnificus.
TO BE PUBLISHED
|
|
3T4P
 
 | |
3ZM6
 
 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | N-(6-(4-(2h-tetrazol-5-yl)benzyl)-3-cyano-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)-2,4-dichloro-5-(morpholinosulfonyl)benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
3ZMM
 
 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-FLUORO-4-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHOXY]-N-(5-METHYL-1H-PYRAZOL-3-YL)-6-MORPHOLINO-PYRIMIDIN-2-AMINE, ACETYL GROUP, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Novel Jak2-Stat Pathway Inhibitors with Extended Residence Time on Target.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5UAD
 
 | |
3ZOQ
 
 | | Structure of BsUDG-p56 complex | | Descriptor: | CHLORIDE ION, GLYCEROL, P56, ... | | Authors: | Banos-Sanz, J.I, Mojardin, L, Sanz-Aparicio, J, Gonzalez, B, Salas, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure and Functional Insights Into Uracil-DNA Glycosylase Inhibition by Phage Phi29 DNA Mimic Protein P56
Nucleic Acids Res., 41, 2013
|
|
3ZDL
 
 | | Vinculin head (1-258) in complex with a RIAM fragment | | Descriptor: | AMYLOID BETA A4 PRECURSOR PROTEIN-BINDING FAMILY B MEMBER 1-INTERACTING PROTEIN, VINCULIN | | Authors: | Zacharchenko, T, Elliott, P.R, Goult, B.T, Bate, N, Critchely, D.R, Barsukov, I.L. | | Deposit date: | 2012-11-28 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Riam and Vinculin Binding to Talin are Mutually Exclusive and Regulate Adhesion Assembly and Turnover.
J.Biol.Chem., 288, 2013
|
|
3SRE
 
 | | Serum paraoxonase-1 by directed evolution at pH 6.5 | | Descriptor: | BROMIDE ION, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ben David, M, Elias, M, Silman, I, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1.
J.Mol.Biol., 418, 2012
|
|
3ZBO
 
 | | A new family of proteins related to the HEAT-like repeat DNA glycosylases with affinity for branched DNA structures | | Descriptor: | ALKF, CHLORIDE ION | | Authors: | Backe, P.H, Simm, R, Laerdahl, J.K, Dalhus, B, Fagerlund, A, Okstad, O.A, Rognes, T, Alseth, I, Kolsto, A.-B, Bjoras, M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A New Family of Proteins Related to the Heat-Like Repeat DNA Glycosylases with Affinity for Branched DNA Structures.
J.Struct.Biol., 183, 2013
|
|
3TEB
 
 | | endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b | | Descriptor: | Endonuclease/exonuclease/phosphatase, MAGNESIUM ION | | Authors: | Chang, C, Bigelow, L, Muniez, I, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-12 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b
To be Published
|
|
4H3P
 
 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Toeroe, I, Remenyi, A. | | Deposit date: | 2012-09-14 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-peptide complex crystallization: a case study on the ERK2 mitogen-activated protein kinase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H6O
 
 | | Sterol 14-alpha demethylase (CYP51)from Trypanosoma cruzi in complex with the inhibitor NEU321 (1-(3-(4-chloro-3,5-dimethylphenoxy)benzyl)-1H-imidazole | | Descriptor: | 1-[3-(4-chloro-3,5-dimethylphenoxy)benzyl]-1H-imidazole, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Wawrzak, Z, Hargrove, T.Y, Pollastri, M.P, Lepesheva, G.I. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antitrypanosomal Lead Discovery: Identification of a Ligand-Efficient Inhibitor of Trypanosoma cruzi CYP51 and Parasite Growth.
J.Med.Chem., 56, 2013
|
|
3S4U
 
 | |
