4HDP
 
 | | Crystal Structure of HIV-1 protease mutants I50V complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Shen, C.H, Zhang, H, Weber, I.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
6HQ2
 
 | | Structure of EAL Enzyme Bd1971 - apo form | | Descriptor: | EAL Enzyme Bd1971 | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
6ISW
 
 | | Structure of human telomeric DNA with 5-selenophene-modified deoxyuridine at residue 12 | | Descriptor: | DNA (22-MER), POTASSIUM ION | | Authors: | Saikrishnan, K, Nuthanakanti, A, Srivatsan, S.G, Ahmad, I. | | Deposit date: | 2018-11-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing G-quadruplex topologies and recognition concurrently in real time and 3D using a dual-app nucleoside probe.
Nucleic Acids Res., 47, 2019
|
|
6HQF
 
 | | Structure of Phenylalanine ammonia-lyase from Petroselinum crispum in complex with (R)-APEP | | Descriptor: | Phenylalanine ammonia-lyase 1, [(1R)-1-amino-2-phenylethyl]phosphonic acid | | Authors: | Bata, Z, Molnar, B, Leveles, I, Poppe, L, Vertessy, G.B. | | Deposit date: | 2018-09-24 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Substrate Tunnel Engineering Aided by X-ray Crystallography and Functional Dynamics Swaps the Function of MIO-Enzymes
Acs Catalysis, 2021
|
|
4K3H
 
 | | Immunoglobulin lambda variable domain L5(L89S) fluorogen activationg protein in complex with malachite green | | Descriptor: | 4-{bis[4-(dimethylamino)phenyl]methyl}phenol, GLYCEROL, Immunoglobulin lambda variable domain L5(L89S), ... | | Authors: | Stanfield, R.L, Szent-Gyorgyi, C, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Malachite Green Mediates Homodimerization of Antibody VL Domains to Form a Fluorescent Ternary Complex with Singular Symmetric Interfaces.
J.Mol.Biol., 425, 2013
|
|
4GSL
 
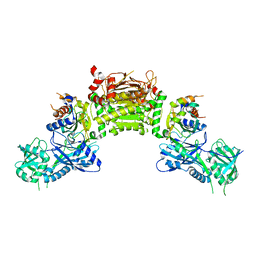 | | Crystal structure of an Atg7-Atg3 crosslinked complex | | Descriptor: | Autophagy-related protein 3, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Kaiser, S.E, Mao, K, Taherbhoy, A.M, Yu, S, Olszewski, J.L, Duda, D.M, Kurinov, I, Deng, A, Fenn, T.D, Klionsky, D.J, Schulman, B.A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1USD
 
 | | human VASP tetramerisation domain L352M | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Kuhnel, K, Jarchau, T, Wolf, E, Schlichting, I, Walter, U, Wittinghofer, A, Strelkov, S.V. | | Deposit date: | 2003-11-21 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Vasp Tetramerization Domain is a Right-Handed Coiled Coil Based on a 15-Residue Repeat
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SEG
 
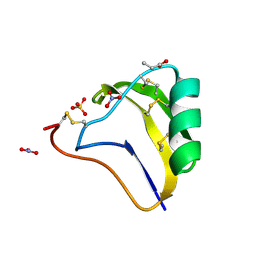 | | Crystal structure of a toxin chimera between Lqh-alpha-IT from the scorpion Leiurus quinquestriatus hebraeus and AAH2 from Androctonus australis hector | | Descriptor: | AAH2: LQH-ALPHA-IT (FACE) CHIMERIC TOXIN, NITRATE ION, PROPANOIC ACID, ... | | Authors: | Karbat, I, Frolow, F, Froy, O, Gilles, N, Cohen, L, Turkov, M, Gordon, D, Gurevitz, M. | | Deposit date: | 2004-02-17 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of the high insecticidal potency of scorpion alpha-toxins.
J.Biol.Chem., 279, 2004
|
|
1USE
 
 | | human VASP tetramerisation domain | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Kuhnel, K, Jarchau, T, Wolf, E, Schlichting, I, Walter, U, Wittinghofer, A, Strelkov, S.V. | | Deposit date: | 2003-11-21 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Vasp Tetramerization Domain is a Right-Handed Coiled Coil Based on a 15-Residue Repeat
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UUD
 
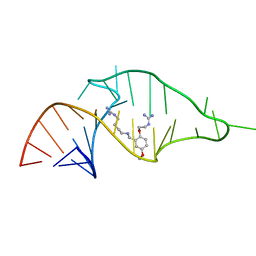 | | NMR structure of a synthetic small molecule, rbt203, bound to HIV-1 TAR RNA | | Descriptor: | N-[2-(2-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-4-METHOXYPHENOXY)ETHYL]GUANIDINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
3NUO
 
 | |
4GXQ
 
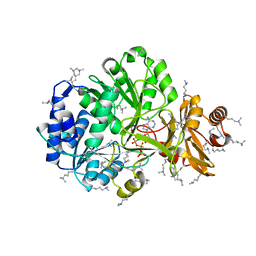 | | Crystal Structure of ATP bound RpMatB-BxBclM chimera B1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Substrate Specificity of the Rhodopseudomonas palustris Protein Acetyltransferase RpPat: IDENTIFICATION OF A LOOP CRITICAL FOR RECOGNITION BY RpPat.
J.Biol.Chem., 287, 2012
|
|
4K90
 
 | | Extracellular metalloproteinase from Aspergillus | | Descriptor: | BORIC ACID, CALCIUM ION, Extracellular metalloproteinase mep, ... | | Authors: | Fernandez, D, Russi, S, Vendrell, J, Monod, M, Pallares, I. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A functional and structural study of the major metalloprotease secreted by the pathogenic fungus Aspergillus fumigatus.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HE9
 
 | | Crystal Structure of HIV-1 protease mutants I54M complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 protease, ... | | Authors: | Shen, C.H, Zhang, H, Weber, I.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
5LOF
 
 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
6IP7
 
 | | Structure of human telomeric DNA with 5-Selenophene-modified deoxyuridine at residue 11 | | Descriptor: | DNA (22-MER), POTASSIUM ION | | Authors: | Saikrishnan, K, Nuthanakanti, A, Srivatsan, S.G, Ahmad, I. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing G-quadruplex topologies and recognition concurrently in real time and 3D using a dual-app nucleoside probe.
Nucleic Acids Res., 47, 2019
|
|
5WBA
 
 | | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins - WT | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
6HN9
 
 | | Nicomicin-1 -- Novel antimicrobial peptides from the Arctic polychaeta Nicomache minor provide new molecular insight into biological role of the BRICHOS domain | | Descriptor: | Nicomicin-1 | | Authors: | Panteleev, P.V, Tsarev, A.V, Bolosov, I.A, Paramonov, A.S, Marggraf, M.B, Sychev, S.V, Shenkarev, Z.O, Ovchinnikova, T.V. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Antimicrobial Peptides from the Arctic PolychaetaNicomache minorProvide New Molecular Insight into Biological Role of the BRICHOS Domain.
Mar Drugs, 16, 2018
|
|
6HYK
 
 | | NMR solution structure of the C/D box snoRNA U14 | | Descriptor: | RNA (31-MER) | | Authors: | Chagot, M.E, Quinternet, M, Rothe, B, Charpentier, B, Coutant, J, Manival, X, Lebars, I. | | Deposit date: | 2018-10-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The yeast C/D box snoRNA U14 adopts a "weak" K-turn like conformation recognized by the Snu13 core protein in solution.
Biochimie, 164, 2019
|
|
3NU9
 
 | |
8A4Y
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with N-(2,3-dihydro-1H-inden-5-yl)acetamide | | Descriptor: | Host translation inhibitor nsp1, N-(2,3-dihydro-1H-inden-5-yl)acetamide | | Authors: | Borsatto, A, Galdadas, I, Ma, S, Damfo, S, Haider, S, Kozielski, F, Estarellas, C, Gervasio, F.L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Revealing druggable cryptic pockets in the Nsp1 of SARS-CoV-2 and other beta-coronaviruses by simulations and crystallography.
Elife, 11, 2022
|
|
4L28
 
 | | Crystal structure of delta516-525 human cystathionine beta-synthase D444N mutant containing C-terminal 6xHis tag | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez, L.A. | | Deposit date: | 2013-06-04 | | Release date: | 2013-09-18 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8A48
 
 | | Less crystallisable" IgG1 Fc fragment (E382S variant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc, ... | | Authors: | Sudol, A.S.L, Tews, I, Crispin, M. | | Deposit date: | 2022-06-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.044 Å) | | Cite: | Extensive substrate recognition by the streptococcal antibody-degrading enzymes IdeS and EndoS.
Nat Commun, 13, 2022
|
|
6I6M
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
1Y6N
 
 | | Crystal structure of Epstein-Barr virus IL-10 mutant (A87I) complexed with the soluble IL-10R1 chain | | Descriptor: | Interleukin-10 receptor alpha chain, Viral interleukin-10 homolog | | Authors: | Yoon, S.I, Jones, B.C, Logsdon, N.J, Walter, M.R. | | Deposit date: | 2004-12-06 | | Release date: | 2005-05-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain.
Structure, 13, 2005
|
|
