6KUC
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 2) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
3CZK
 
 | | Crystal Structure Analysis of Sucrose hydrolase(SUH) E322Q-sucrose complex | | Descriptor: | Sucrose hydrolase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
1SWL
 
 | | CORE-STREPTAVIDIN MUTANT W108F AT PH 7.0 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWJ
 
 | | CORE-STREPTAVIDIN MUTANT W79F AT PH 4.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWO
 
 | | CORE-STREPTAVIDIN MUTANT W120F AT PH 7.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWE
 
 | | APO-CORE-STREPTAVIDIN IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWD
 
 | | APO-CORE-STREPTAVIDIN IN COMPLEX WITH BIOTIN (TWO UNOCCUPIED BINDING SITES) AT PH 4.5 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
4RNJ
 
 | | PaMorA phosphodiesterase domain, apo form | | Descriptor: | Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
3FNM
 
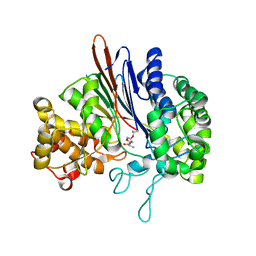 | | Crystal structure of acivicin-inhibited gamma-glutamyltranspeptidase reveals critical roles for its C-terminus in autoprocessing and catalysis | | Descriptor: | (2S)-AMINO[(5S)-3-CHLORO-4,5-DIHYDROISOXAZOL-5-YL]ACETIC ACID, Gamma-glutamyltranspeptidase (Ggt) Large subunit, Gamma-glutamyltranspeptidase (Ggt) Small subunit | | Authors: | Williams, K, Cullati, S, Sand, A, Biterova, E.I, Barycki, J.J. | | Deposit date: | 2008-12-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Acivicin-Inhibited gamma-Glutamyltranspeptidase Reveals Critical Roles for Its C-Terminus in Autoprocessing and Catalysis.
Biochemistry, 48, 2009
|
|
1XRF
 
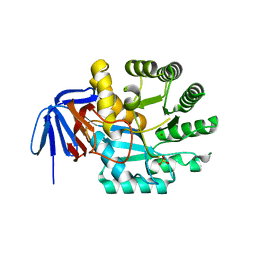 | | The Crystal Structure of a Novel, Latent Dihydroorotase from Aquifex aeolicus at 1.7 A resolution | | Descriptor: | Dihydroorotase, SULFATE ION, ZINC ION | | Authors: | Martin, P.D, Purcarea, C, Zhang, P, Vaishnav, A, Sadecki, S, Guy-Evans, H.I, Evans, D.R, Edwards, B.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a novel, latent dihydroorotase from Aquifex aeolicus at 1.7A resolution
J.Mol.Biol., 348, 2005
|
|
4RNH
 
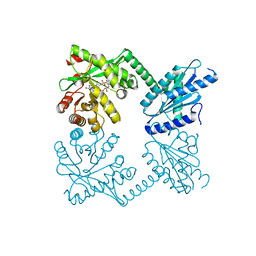 | | PaMorA tandem diguanylate cyclase - phosphodiesterase, c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
1SWF
 
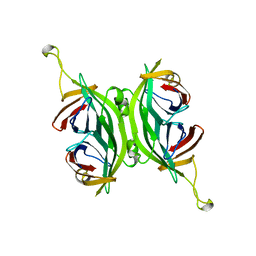 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 | | Descriptor: | CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-04-23 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
3CKW
 
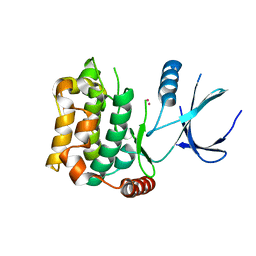 | | Crystal structure of sterile 20-like kinase 3 (MST3, STK24) | | Descriptor: | MERCURY (II) ION, Serine/threonine-protein kinase 24 | | Authors: | Antonysamy, S.S, Burley, S.K, Buchanan, S, Chau, F, Feil, I, Wu, L, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-17 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of sterile 20-like kinase 3 (MST3, STK24).
To be Published
|
|
4RPE
 
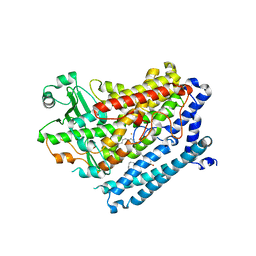 | |
3CMH
 
 | | SYNTHETIC LINEAR TRUNCATED ENDOTHELIN-1 AGONIST | | Descriptor: | PROTEIN (ENDOTHELIN-1) | | Authors: | Hewage, C.M, Jiang, L, Parkinson, J.A, Ramage, R, Sadler, I.H. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel ETB receptor selective agonist ET1-21 [Cys(Acm)1,15, Aib3,11, Leu7] by nuclear magnetic resonance spectroscopy and molecular modelling.
J.Pept.Res., 53, 1999
|
|
1SA1
 
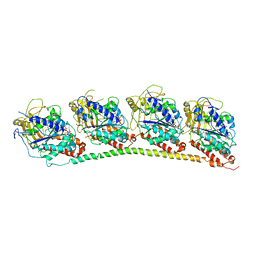 | | Tubulin-podophyllotoxin: stathmin-like domain complex | | Descriptor: | 9-HYDROXY-5-(3,4,5-TRIMETHOXYPHENYL)-5,8,8A,9-TETRAHYDROFURO[3',4':6,7]NAPHTHO[2,3-D][1,3]DIOXOL-6(5AH)-ONE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ravelli, R.B, Gigant, B, Curmi, P.A, Jourdain, I, Lachkar, S, Sobel, A, Knossow, M. | | Deposit date: | 2004-02-06 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain.
Nature, 428, 2004
|
|
4RSX
 
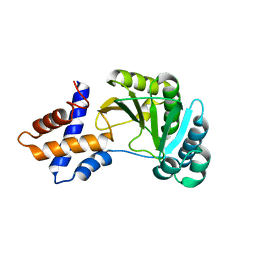 | |
2XNV
 
 | | Acetylcholine binding protein (AChBP) as template for hierarchical in silico screening procedures to identify structurally novel ligands for the nicotinic receptors | | Descriptor: | 2-(2-(4-PHENYLPIPERIDIN-1-YL)ETHYL)-1H-INDOLE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Rucktooa, P, Akdemir, A, deEsch, I, Sixma, T.K. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Acetylcholine Binding Protein (Achbp) as Template for Hierarchical in Silico Screening Procedures to Identify Structurally Novel Ligands for the Nicotinic Receptors.
Bioorg.Med.Chem., 19, 2011
|
|
4N37
 
 | | Structure of langerin CRD I313 D288 complexed with Me-Man | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, methyl alpha-D-mannopyranoside | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
6KSV
 
 | | Crystal structure of SurE with D-Leu | | Descriptor: | Alpha/beta hydrolase, D-LEUCINE, S-(2-acetamidoethyl) (2R)-2-azanyl-4-methyl-pentanethioate, ... | | Authors: | Zhai, R, Mori, T, Abe, I. | | Deposit date: | 2019-08-26 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Heterochiral coupling in non-ribosomal peptide macrolactamization
Nat Catal, 2020
|
|
1XRJ
 
 | | Rapid structure determination of human uridine-cytidine kinase 2 using a conventional laboratory X-ray source and a single samarium derivative | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Appleby, T.C, Larson, G, Wu, J.Z, Cheney, I.W, Hong, Z, Yao, N. | | Deposit date: | 2004-10-14 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human uridine-cytidine kinase 2 determined by SIRAS using a rotating-anode X-ray generator and a single samarium derivative.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2M5C
 
 | |
3D3L
 
 | | The 2.6 A crystal structure of the lipoxygenase domain of human arachidonate 12-lipoxygenase, 12S-type | | Descriptor: | Arachidonate 12-lipoxygenase, 12S-type, FE (III) ION | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Van Den Berg, S, Welin, M, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-12 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the lipoxygenase domain of human Arachidonate 12-lipoxygenase, 12S-type.
To be Published
|
|
2M5D
 
 | |
6KUB
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
