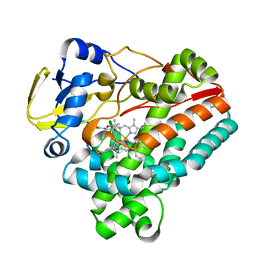
4O5I
 
 | |
5CK5
 
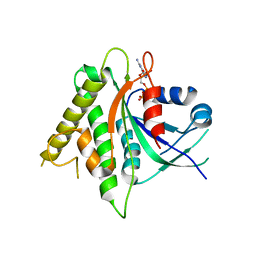 | |
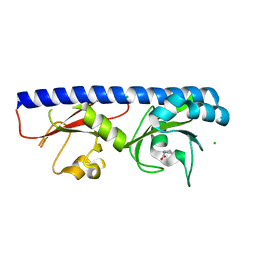
6Q0G
 
 | |
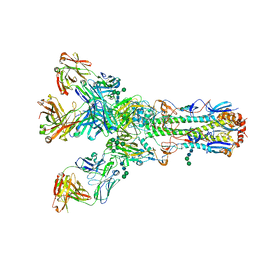
6WLL
 
 | | Apo F. nucleatum glycine riboswitch models, 10.0 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6J84
 
 | | Crystal structure of TleB with hydroxyl analog | | Descriptor: | (2S)-2-hydroxy-N-[(2S)-1-hydroxy-3-(1H-indol-3-yl)propan-2-yl]-3-methylbutanamide, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakamura, H, Mori, T, Abe, I. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
5K23
 
 | |
5GYJ
 
 | | Structure of catalytically active sortase from Clostridium difficile | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Yin, J.-C, Fei, C.-H, Hsiao, Y.-Y, Nix, J.C, Huang, I.-H, Wang, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Insights into Substrate Recognition by Clostridium difficile Sortase.
Front Cell Infect Microbiol, 6, 2016
|
|
6PUW
 
 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and Bictegravir (BIC) | | Descriptor: | Bictegravir, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
2Z4O
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
1SIS
 
 | | SPATIAL STRUCTURE OF INSECTOTOXIN I5A BUTHUS EUPEUS BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY (RUSSIAN) | | Descriptor: | SCORPION INSECTOTOXIN I5A | | Authors: | Arseniev, A.S, Kondakov, V.I, Lomize, A.L, Maiorov, V.N, Bystrov, V.F. | | Deposit date: | 1993-11-11 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the spatial structure of insectotoxin 15A from Buthus erpeus by (1)H-NMR spectroscopy data
Bioorg.Khim., 17, 1991
|
|
1SJB
 
 | | X-ray structure of o-succinylbenzoate synthase complexed with o-succinylbenzoic acid | | Descriptor: | 2-SUCCINYLBENZOATE, MAGNESIUM ION, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
6T46
 
 | | Structure of the Rap conjugation gene regulator of the plasmid pLS20 in complex with the Phr* peptide | | Descriptor: | CHLORIDE ION, Quorum-sensing secretion protein (processed), Response regulator aspartate phosphatase, ... | | Authors: | Crespo, I, Bernardo, N, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2019-10-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inactivation of the dimeric RappLS20 anti-repressor of the conjugation operon is mediated by peptide-induced tetramerization.
Nucleic Acids Res., 48, 2020
|
|
3P4U
 
 | | Crystal structure of active caspase-6 in complex with Ac-VEID-CHO inhibitor | | Descriptor: | Ac-VEID-CHO inhibitor, Caspase-6 | | Authors: | Mueller, I, Lamers, M.B.A.C, Ritchie, A.J, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2010-10-07 | | Release date: | 2011-09-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of active and inhibitor-bound human Casp6
To be Published
|
|
3P68
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | GOLD 3+ ION, Lysozyme C | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
6WLT
 
 | | Apo V. cholerae glycine riboswitch models, 4.8 Angstrom resolution | | Descriptor: | RNA (231-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
2BWP
 
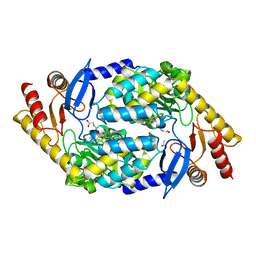 | | 5-Aminolevulinate Synthase from Rhodobacter capsulatus in complex with glycine | | Descriptor: | 5-AMINOLEVULINATE SYNTHASE, ACETIC ACID, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Astner, I, Schulze, J.O, Van Den Heuvel, J.J, Jahn, D, Schubert, W.-D, Heinz, D.W. | | Deposit date: | 2005-07-15 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of 5-Aminolevulinate Synthase, the First Enzyme of Heme Biosynthesis, and its Link to Xlsa in Humans.
Embo J., 24, 2005
|
|
6PUZ
 
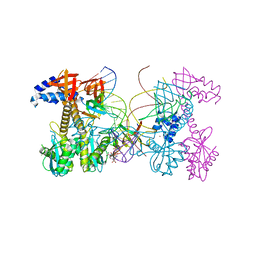 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6Q9T
 
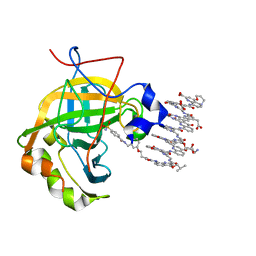 | | Protein-aromatic foldamer complex crystal structure | | Descriptor: | 2-(8-azanyl-2-methanoyl-quinolin-4-yl)ethanoic acid, 6-(aminomethyl)pyridine-2-carboxylic acid, 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, ... | | Authors: | Post, S, Langlois d'Estaintot, B, Fischer, L, Granier, T, Huc, I. | | Deposit date: | 2018-12-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure Elucidation of Helical Aromatic Foldamer-Protein Complexes with Large Contact Surface Areas.
Chemistry, 25, 2019
|
|
6PXY
 
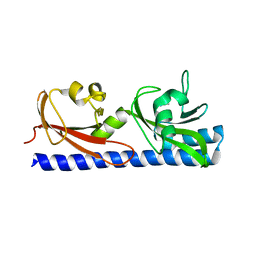 | |
1SOM
 
 | | TORPEDO CALIFORNICA ACETYLCHOLINESTERASE INHIBITED BY NERVE AGENT GD (SOMAN). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METHYLPHOSPHONIC ACID ESTER GROUP, PROTEIN (ACETYLCHOLINESTERASE), ... | | Authors: | Greenblatt, H.M, Millard, C.B, Sussman, J.L, Silman, I. | | Deposit date: | 1999-03-17 | | Release date: | 1999-06-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level.
Biochemistry, 38, 1999
|
|
7WP3
 
 | | Crystal structure of the complex of proliferating cell nuclear antigen (PCNA) from Leishmania donovani with 1,5-Bis (4-amidinophenoxy) pentane (PNT) at 2.95 A resolution | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, Proliferating cell nuclear antigen | | Authors: | Ahmad, M.I, Yadav, S.P, Singh, P.K, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-01-22 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Crystal structure of the complex of proliferating cell nuclear antigen (PCNA) from Leishmania donovani with 1,5-Bis (4-amidinophenoxy) pentane (PNT) at 2.95 A resolution
To Be Published
|
|
5CJH
 
 | | Crystal Structure of Eukaryotic Oxoiron MagKatG2 at pH 8.5 | | Descriptor: | Catalase-peroxidase 2, HYDROXIDE ION, Peroxidized Heme | | Authors: | Gasselhuber, B, Obinger, C, Fita, I, Carpena, X. | | Deposit date: | 2015-07-14 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Eukaryotic Catalase-Peroxidase: The Role of the Trp-Tyr-Met Adduct in Protein Stability, Substrate Accessibility, and Catalysis of Hydrogen Peroxide Dismutation.
Biochemistry, 54, 2015
|
|
6X1V
 
 | | Structure of pHis Fab (SC44-8) in complex with pHis mimetic peptide | | Descriptor: | 1,2-ETHANEDIOL, ACLYana-3-pTza peptide, SC44-8 Heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6QF7
 
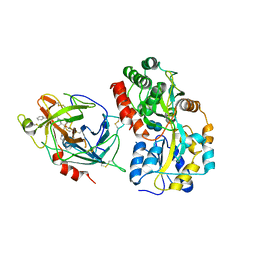 | | Crystal structures of the recombinant beta-Factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, ... | | Authors: | Pathak, M, Mannal, R, Li, C, Bubacarr, G.K, Badraldin, K.H, Belviso, B.D, Camila, R.B, Dreveny, I, Fischer, P.M, Dekker, L.V, Oliva, M.L.V, Emsley, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structures of the recombinant beta-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6WIL
 
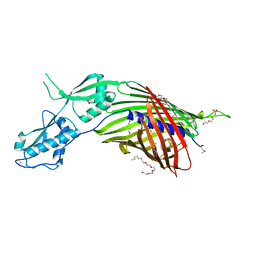 | | CdiB from Acinetobacter baumannii | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, DI(HYDROXYETHYL)ETHER, Hemolysin activator protein CdiB, ... | | Authors: | Guerin, J, Botos, I, Buchanan, S.K. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into toxin secretion by contact dependent growth inhibition transporters.
Elife, 9, 2020
|
|
