1Q0K
 
 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the thiosulfate-reduced state | | Descriptor: | NICKEL (II) ION, SULFATE ION, Superoxide dismutase [Ni], ... | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8RDJ
 
 | | Plastid-encoded RNA polymerase transcription elongation complex (Integrated model) | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
7UQ6
 
 | | tRNA T-box antiterminator fusion, construct #4 | | Descriptor: | BARIUM ION, POTASSIUM ION, RNA (86-MER) | | Authors: | Grigg, J.C, Price, I.R, Ke, A. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | tRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-tRNA Sensing Mechanisms by the T-Box Riboswitch
Crystals, 12, 2022
|
|
1I79
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND 5'-DEOXY-5'-[(3-HYDRAZINOPROPYL)METHYLAMINO]ADENOSINE | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-[(3-HYDRAZINOPROPYL)METHYLAMINO]ADENOSINE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
4GEX
 
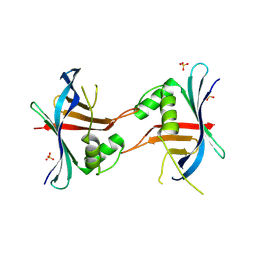 | |
8RAS
 
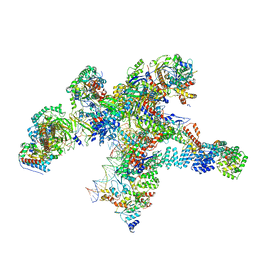 | | Plastid-encoded RNA polymerase transcription elongation complex | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
4YHP
 
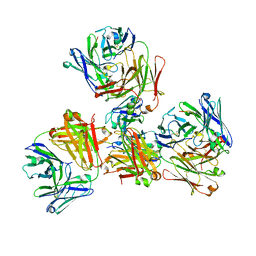 | | Crystal structure of 309M3-B Fab in complex with H3K9me3 peptide | | Descriptor: | Fab Heavy Chain, Fab Light Chain, H3K9me3 peptide | | Authors: | Hattori, T, Dementieva, I.S, Montano, S.P, Koide, S. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2FX9
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with a thioether-linked peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein gp41 | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
8BNY
 
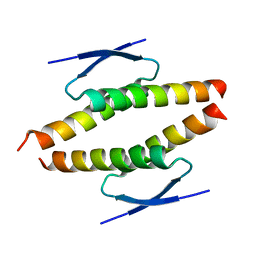 | | Structure of the tetramerization domain of pLS20 conjugation repressor Rco | | Descriptor: | CHLORIDE ION, Immunity repressor protein | | Authors: | Bernardo, N, Crespo, I, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | A tetramerization domain in prokaryotic and eukaryotic transcription regulators homologous to p53.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1LII
 
 | | STRUCTURE OF T. GONDII ADENOSINE KINASE BOUND TO ADENOSINE 2 AND AMP-PCP | | Descriptor: | ADENOSINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Scott, D.M, Mathews, I.I, Ealick, S.E, Roos, D.S, Ullman, B, Brennan, R.G. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding.
J.Mol.Biol., 298, 2000
|
|
8B43
 
 | | Crystal structure of ferrioxamine transporter | | Descriptor: | 1,12-bis(oxidanyl)-1,6,12,17-tetrazacyclodocosane-2,5,13,16-tetrone, FE (III) ION, Ferrichrome-iron receptor, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Interactions of TonB-dependent transporter FoxA with siderophores and antibiotics that affect binding, uptake, and signal transduction.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BI3
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200W (crystal M200W#1) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8BKF
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (crystal M200T#o) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-09 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.221 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8BP9
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200W (crystal M200W#2) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8UD7
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8SDG
 
 | |
3MDY
 
 | | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Bone morphogenetic protein receptor type-1B, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chaikuad, A, Sanvitale, C, Mahajan, P, Daga, N, Cooper, C, Krojer, T, Alfano, I, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189
To be Published
|
|
8SDF
 
 | |
8SR6
 
 | |
8BS8
 
 | | Bovine naive ultralong antibody AbD08 collected at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heavy chain, Light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The impact of exchanging the light and heavy chains on the structures of bovine ultralong antibodies.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8BQO
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant M200I | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoaspartyl peptidase subunit alpha, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
5JJF
 
 | | Structure of the SRII/HtrII Complex in I212121 space group ("U" shape) - M state | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-23 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
7VFL
 
 | | Crystal structure of SdgB (UDP, NAG, and O-glycosylated SD peptide-binding form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFO
 
 | | Crystal structure of SdgB (Phosphate-binding form) | | Descriptor: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFK
 
 | | Crystal structure of SdgB (ligand-free form) | | Descriptor: | GLYCEROL, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
