2AQW
 
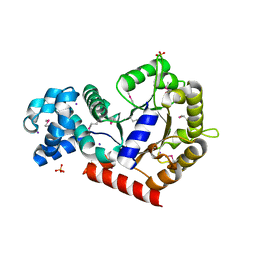 | | Structure of putative orotidine-monophosphate-decarboxylase from Plasmodium yoelii (PY01515) | | Descriptor: | IODIDE ION, SULFATE ION, putative orotidine-monophosphate-decarboxylase | | Authors: | Dong, A, Vedadi, M, Wasney, G, Zhao, Y, Lew, J, Alam, Z, Melone, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
8T9L
 
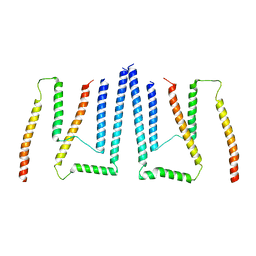 | | Pom34-Pom152 membrane attachment site yeast NPC | | Descriptor: | Nucleoporin POM152, Nucleoporin POM34 | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-06-24 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
1QCZ
 
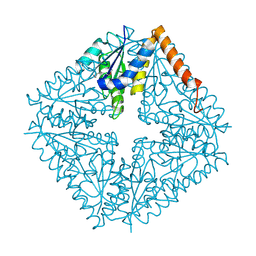 | |
6IOU
 
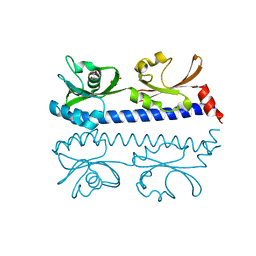 | | The ligand binding domain of Mlp24 with serine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
5U3L
 
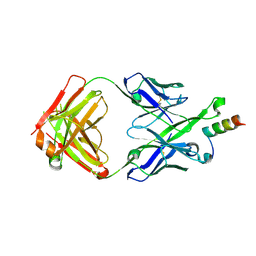 | | Crystal Structure of DH511.2 Fab in Complex with HIV-1 gp41 MPER 670-683 Peptide | | Descriptor: | DH511.2 Fab Heavy Chain, DH511.2 Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
8Q9X
 
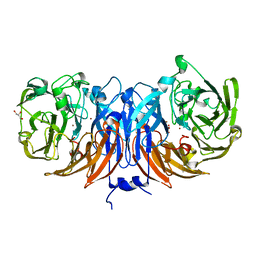 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum with molecular oxygen at 1.05 A resolution | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-08-22 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
8Q9Y
 
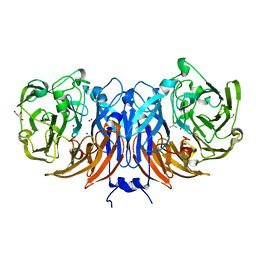 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum in complex with inhibitor thiourea at 1.10 A resolution | | Descriptor: | COPPER (II) ION, GLYCEROL, THIOUREA, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-08-22 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
4HXG
 
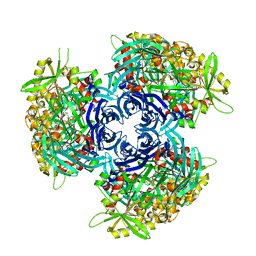 | | Pyrococcus horikoshii acylaminoacyl peptidase (orthorhombic crystal form) | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, MAGNESIUM ION, ... | | Authors: | Kiss-Szeman, A, Menyhard, D.K, Tichy-Racs, E, Hornung, B, Radi, K, Szeltner, Z, Domokos, K, Szamosi, I, Naray-Szabo, G, Polgar, L, Harmat, V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Self-compartmentalizing Hexamer Serine Protease from Pyrococcus Horikoshii: SUBSTRATE SELECTION ACHIEVED THROUGH MULTIMERIZATION.
J.Biol.Chem., 288, 2013
|
|
2KMV
 
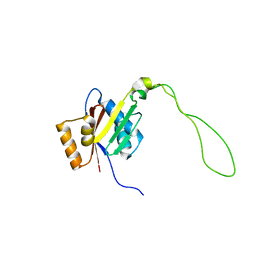 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-free form | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
8PNW
 
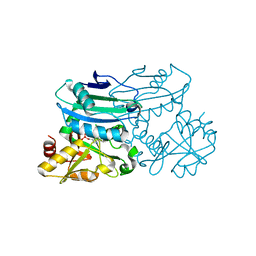 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
7B8E
 
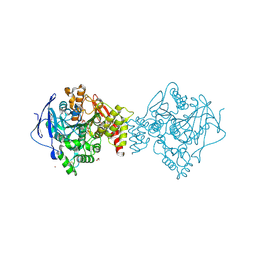 | | Torpedo californica acetylcholinesterase complexed with Ca+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L. | | Deposit date: | 2020-12-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
8QHM
 
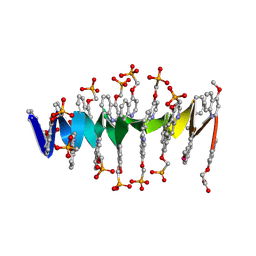 | | DNA mimic Foldamer with sticky ends | | Descriptor: | DNA mimic Foldamer | | Authors: | Deepak, D, Loos, M, Huc, I. | | Deposit date: | 2023-09-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhancing the Features of DNA Mimic Foldamers for Structural Investigations.
Chemistry, 30, 2024
|
|
8PNY
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens complexed with phenylhydrazine and in its apo form | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8T8I
 
 | | Structure of VHH-Fab complex with engineered Elbow FNQIKG, Crystal Kappa and SER substitutions | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab heavy chain, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
1D0I
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR COMPLEXED WITH PHOSPHATE IONS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, TYPE II 3-DEHYDROQUINATE HYDRATASE | | Authors: | Roszak, A.W, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1999-09-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and mechanism of the type II dehydroquinase from Streptomyces coelicolor.
Structure, 10, 2002
|
|
7KQR
 
 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | Authors: | Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
2CIV
 
 | | Chloroperoxidase bromide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CHLOROPEROXIDASE, ... | | Authors: | Kuhnel, K, Blankenfeldt, W, Terner, J, Schlichting, I. | | Deposit date: | 2006-03-26 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Chloroperoxidase with its Bound Substrates and Complexed with Formate, Acetate, and Nitrate.
J.Biol.Chem., 281, 2006
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
2LB7
 
 | | Hevein-type Antifungal Peptide with a Unique 10-Cysteine Motif | | Descriptor: | Antimicrobial peptide 1a | | Authors: | Balashova, T.A, Vassilevski, A.A, Odintsova, T.I, Grishin, E.V, Egorov, T.A, Arseniev, A.S. | | Deposit date: | 2011-03-23 | | Release date: | 2011-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a defense peptide from wheat with a 10-cysteine motif.
Biochem.Biophys.Res.Commun., 411, 2011
|
|
2L0Y
 
 | | Complex hMia40-hCox17 | | Descriptor: | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) pseudogene (COX17), Mitochondrial intermembrane space import and assembly protein 40 | | Authors: | Bertini, I, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-11-24 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7T1K
 
 | | Crystal structure of a superbinder Fes SH2 domain (sFes1) in complex with a high affinity phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
8CBE
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-2 and Beta-49 Fabs | | Descriptor: | BA.4/5-2 heavy chain, BA.4/5-2 light chain, Beta-49 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
7T1L
 
 | | Crystal structure of a superbinder Fes SH2 domain (sFesS) in complex with a high affinity phosphopeptide | | Descriptor: | CHLORIDE ION, SODIUM ION, Synthetic phosphotyrosine-containing Ezrin-derived peptide, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1U
 
 | | Crystal structure of a superbinder Src SH2 domain (sSrcF) in complex with a high affinity phosphopeptide | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, Synthetic phosphopeptide, ZINC ION | | Authors: | Martyn, G.D, Singer, A.U, Manczyk, N, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
