6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6I4J
 
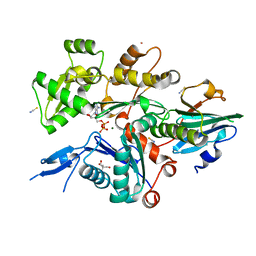 | | Crystal Structure of Plasmodium falciparum actin I (F54Y mutant) in the Mg-ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6I4L
 
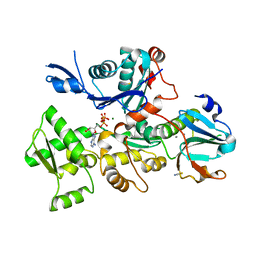 | | Crystal Structure of Plasmodium falciparum actin I (G115A mutant) in the Mg-K-ATP/ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6NQJ
 
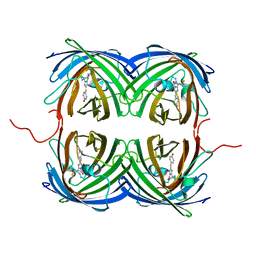 | |
3N03
 
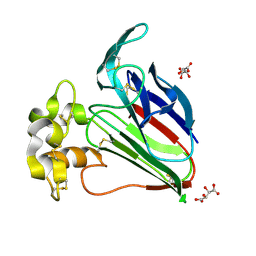 | | Thaumatin crystals grown from drops | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Mathews, I.I. | | Deposit date: | 2010-05-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diffraction study of protein crystals grown in cryoloops and micromounts.
J.Appl.Crystallogr., 43, 2010
|
|
6XC3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.1 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
8F8O
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CITRIC ACID, SUCCINIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids
To Be Published
|
|
7BQ1
 
 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin)-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, PALMITIC ACID, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
4Z2C
 
 | | Quinolone(Moxifloxacin)-DNA cleavage complex of gyrase from S. pneumoniae | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
6I4F
 
 | | Crystal Structure of Plasmodium falciparum actin I (A272W mutant) in the Mg-K-ATP/ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6T2Q
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16, ... | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
3K2S
 
 | | Solution structure of double super helix model | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Apolipoprotein A-I, CHOLESTEROL | | Authors: | Wu, Z, Gogonea, V, Lee, X, Wagner, M.A, Li, X.-M, Huang, Y, Undurti, A, May, R.P, Haertlein, M, Moulin, M, Gutsche, I, Zaccai, G, Didonato, J.A, Hazen, L.S. | | Deposit date: | 2009-09-30 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Double superhelix model of high density lipoprotein.
J.Biol.Chem., 284, 2009
|
|
4Z53
 
 | | Quinolone(Trovafloxacin)-DNA cleavage complex of topoisomerase IV from S. pneumoniae | | Descriptor: | DNA topoisomerase 4 subunit B,DNA topoisomerase 4 subunit A, E-site DNA, MAGNESIUM ION, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-04-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
5OPC
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc and Vadadustat | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Leissing, T.M, Schofield, C.J, Clifton, I.J, Lu, X, Zhang, D. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
5NCE
 
 | | Structure of PsDef1 defensin from Pinus sylvestris | | Descriptor: | Defensin-1 | | Authors: | Khairutdinov, B.I, Ermakova, E.A, Bessolitsyna, E.K, Toporkova, Y.Y, Tarasova, N.B, Kovaleva, V, Zuev, Y.F, Nesmelova, I.V. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure, conformational dynamics, and biological activity of PsDef1 defensin from Pinus sylvestris.
Biochim. Biophys. Acta, 1865, 2017
|
|
6N5M
 
 | |
1RXS
 
 | | E. coli uridine phosphorylase: 2'-deoxyuridine phosphate complex | | Descriptor: | 2'-DEOXYURIDINE, META VANADATE, PHOSPHATE ION, ... | | Authors: | Caradoc-Davies, T.T, Cutfield, S.M, Lamont, I.L, Cutfield, J.F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium
J.Mol.Biol., 337, 2004
|
|
3MTF
 
 | | Crystal structure of the ACVR1 kinase in complex with a 2-aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Canning, P, Krojer, T, Vollmar, M, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
3MX9
 
 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*GP*CP*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*GP*AP*GP*AP*AP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*TP*CP*TP*CP*AP*GP*GP*TP*AP*CP*CP*TP*CP*AP*GP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Montoya, G. | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus.
Nucleic Acids Res., 39, 2011
|
|
6QKZ
 
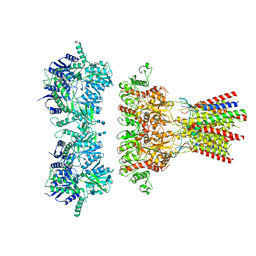 | | Full length GluA1/2-gamma8 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I.G. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma 8.
Science, 364, 2019
|
|
6HCW
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Cryptosporidium parvum complexed with L-lysine and a difluoro cyclohexyl chromone ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7BQ2
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BVX
 
 | | Crystal structure of C-terminal fragment of pilus adhesin SpaC from Lactobacillus rhamnosus GG-Iodide soaked | | Descriptor: | IODIDE ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-04-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
1ZYM
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI | | Descriptor: | ENZYME I | | Authors: | Liao, D.-I, Davies, D.R. | | Deposit date: | 1996-05-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The first step in sugar transport: crystal structure of the amino terminal domain of enzyme I of the E. coli PEP: sugar phosphotransferase system and a model of the phosphotransfer complex with HPr.
Structure, 4, 1996
|
|
8QU2
 
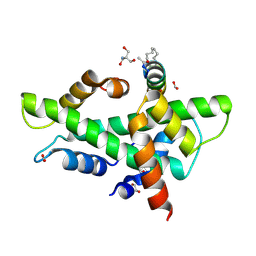 | | NF-YB/C Heterodimer in Complex with a 16-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Durukan, C, Arbore, F, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
