1R02
 
 | |
1Z7G
 
 | | Free human HGPRT | | 分子名称: | Hypoxanthine-guanine phosphoribosyltransferase | | 著者 | Keough, D.T, Brereton, I.M, de Jersey, J, Guddat, L.W. | | 登録日 | 2005-03-24 | | 公開日 | 2005-08-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Crystal Structure of Free Human Hypoxanthine-guanine Phosphoribosyltransferase Reveals Extensive Conformational Plasticity Throughout the Catalytic Cycle
J.Mol.Biol., 351, 2005
|
|
1QU5
 
 | |
1QWL
 
 | | Structure of Helicobacter pylori catalase | | 分子名称: | AZIDE ION, KatA catalase, OXYGEN MOLECULE, ... | | 著者 | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Obenbreit, S, Nicholls, P, Fita, I. | | 登録日 | 2003-09-02 | | 公開日 | 2004-03-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
1ZPC
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 2-[2-(3-Chloro-phenyl)-2-hydroxy-acetylamino]-N-[4-guanidino-1-(thiazole-2-carbonyl)-butyl]-3-methyl-butyramide | | 分子名称: | 2-[2-(3-CHLORO-PHENYL)-2-HYDROXY-ACETYLAMINO]-N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-3-METHYL-BUTYRAMIDE, Coagulation factor XI, SULFATE ION | | 著者 | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | 登録日 | 2005-05-16 | | 公開日 | 2006-04-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1QVB
 
 | | CRYSTAL STRUCTURE OF THE BETA-GLYCOSIDASE FROM THE HYPERTHERMOPHILE THERMOSPHAERA AGGREGANS | | 分子名称: | BETA-GLYCOSIDASE | | 著者 | Chi, Y.-I, Martinez-Cruz, L.A, Swanson, R.V, Robertson, D.E, Kim, S.-H. | | 登録日 | 1999-07-07 | | 公開日 | 1999-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the beta-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability.
FEBS Lett., 445, 1999
|
|
1QW6
 
 | | Rat neuronal nitric oxide synthase oxygenase domain in complex with N-omega-propyl-L-Arg. | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-PROPYL-L-ARGININE, Nitric-oxide synthase, ... | | 著者 | Fedorov, R, Hartmann, E, Ghosh, D.K, Schlichting, I. | | 登録日 | 2003-09-01 | | 公開日 | 2003-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nomega-propyl-L-Arg for the inducible and neuronal isoforms.
J.Biol.Chem., 278, 2003
|
|
1Z0W
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain at 1.2A resolution | | 分子名称: | CALCIUM ION, Putative protease La homolog type | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | 登録日 | 2005-03-02 | | 公開日 | 2005-08-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
3TWI
 
 | | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores | | 分子名称: | BclA protein, GLYCEROL, Variable lymphocyte receptor B | | 著者 | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | 登録日 | 2011-09-21 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
1QZX
 
 | |
1QWC
 
 | | Rat neuronal nitric oxide synthase oxygenase domain in complex with W1400 inhibitor. | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-(3-(AMINOMETHYL)BENZYL)ACETAMIDINE, Nitric-oxide synthase, ... | | 著者 | Fedorov, R, Hartmann, E, Ghosh, D.K, Schlichting, I. | | 登録日 | 2003-09-02 | | 公開日 | 2003-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nomega-propyl-L-Arg for the inducible and neuronal isoforms.
J.Biol.Chem., 278, 2003
|
|
2A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | 分子名称: | TUMOR SUPPRESSOR P16INK4A | | 著者 | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | 登録日 | 1998-02-13 | | 公開日 | 1999-08-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
3UDN
 
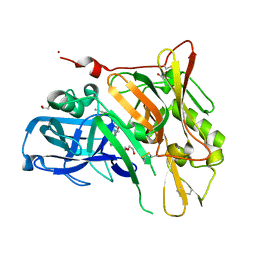 | | Crystal Structure of BACE with Compound 9 | | 分子名称: | 1,2-ETHANEDIOL, 4-cyanobenzyl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, Beta-secretase 1, ... | | 著者 | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | 登録日 | 2011-10-28 | | 公開日 | 2012-04-18 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UDK
 
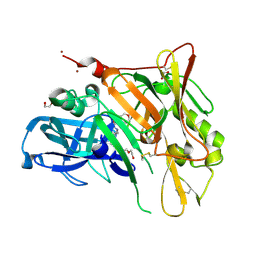 | | Crystal Structure of BACE with Compound 6 | | 分子名称: | 1,2-ETHANEDIOL, Beta-secretase 1, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | 登録日 | 2011-10-28 | | 公開日 | 2012-04-18 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
6KAZ
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by soaking | | 分子名称: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KBA
 
 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by co-crystallization | | 分子名称: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
4XA7
 
 | | Soluble part of holo NqrC from V. harveyi | | 分子名称: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit C | | 著者 | Borshchevskiy, V, Round, E, Bertsova, Y, Polovinkin, V, Gushchin, I, Mishin, A, Kovalev, K, Kachalova, G, Popov, A, Bogachev, A, Gordeliy, V. | | 登録日 | 2014-12-12 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structural and Functional Investigation of Flavin Binding Center of the NqrC Subunit of Sodium-Translocating NADH:Quinone Oxidoreductase from Vibrio harveyi.
Plos One, 10, 2015
|
|
6KJN
 
 | | The microtubule-binding domains of yeast cytoplasmic dynein in the high affinity state | | 分子名称: | Dynein heavy chain, cytoplasmic | | 著者 | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | 登録日 | 2019-07-22 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
4XE4
 
 | | Coagulation Factor XII protease domain crystal structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, GLYCEROL | | 著者 | Pathak, M, Wilmann, P, Awford, J, Li, C, Fisher, P.M, Dreveny, I, Dekker, L.V, Emsley, J. | | 登録日 | 2014-12-22 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Coagulation factor XII protease domain crystal structure.
J.Thromb.Haemost., 13, 2015
|
|
6KAX
 
 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin) co-crystals obtained by cross-seeding | | 分子名称: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6K80
 
 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Complex with CoA and Tryptophol | | 分子名称: | 2-(1H-indol-3-yl)ethanol, ACETYL COENZYME *A, Dopamine N-acetyltransferase | | 著者 | Wu, C.Y, Hu, I.C, Yang, Y.C, Cheng, H.C, Lyu, P.C. | | 登録日 | 2019-06-11 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
4XG2
 
 | | Crystal structure of ligand-free Syk | | 分子名称: | Tyrosine-protein kinase SYK | | 著者 | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | 登録日 | 2014-12-30 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
6QZ7
 
 | | Structure of MBP-Mcl-1 in complex with compound 8b | | 分子名称: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZB
 
 | | Structure of Mcl-1 in complex with compound 8d | | 分子名称: | (2~{R})-2-[[6-ethyl-5-(2-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
4XG8
 
 | | Crystal structure of an inhibitor-bound Syk | | 分子名称: | 1-[(1-{2-[(3-chloro-1-methyl-1H-indazol-5-yl)amino]pyrimidin-4-yl}-3-methyl-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | 著者 | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | 登録日 | 2014-12-30 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
