8S2U
 
 | |
5FEH
 
 | | Crystal structure of PCT64_35B, a broadly neutralizing anti-HIV antibody | | 分子名称: | 1,2-ETHANEDIOL, PCT64_26 Fab heavy chain, PCT64_26 Fab light chain, ... | | 著者 | Murrell, S, Wilson, I.A. | | 登録日 | 2015-12-17 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | HIV Envelope Glycoform Heterogeneity and Localized Diversity Govern the Initiation and Maturation of a V2 Apex Broadly Neutralizing Antibody Lineage.
Immunity, 47, 2017
|
|
5J23
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with 2'-phospho-ADP-ribose | | 分子名称: | 2-hydroxyacid dehydrogenase, ACETATE ION, CHLORIDE ION, ... | | 著者 | Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Kutner, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2016-03-29 | | 公開日 | 2016-04-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
8SIR
 
 | |
8RXA
 
 | |
4BDI
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | 分子名称: | 1,2-ETHANEDIOL, 1-acetyl-N-(5-methylpyridin-2-yl)piperidine-4-carboxamide, CHLORIDE ION, ... | | 著者 | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | 登録日 | 2012-10-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
8SIS
 
 | |
3MGX
 
 | |
4BDB
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | 分子名称: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | 著者 | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | 登録日 | 2012-10-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
8SIQ
 
 | |
6YQO
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW12 | | 分子名称: | (S)-N1-(4-(2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetamido)phenyl)-N8-hydroxyoctanediamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | 著者 | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-04-17 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
5YDV
 
 | | Regulatory domain of HypT from Salmonella typhimurium complexed with HOCl (HOCl-bound form) | | 分子名称: | Cell density-dependent motility repressor, SULFATE ION, hypochlorous acid | | 著者 | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | 登録日 | 2017-09-14 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YEZ
 
 | | Regulatory domain of HypT M206Q mutant from Salmonella typhimurium | | 分子名称: | Cell density-dependent motility repressor | | 著者 | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | 登録日 | 2017-09-20 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3LZG
 
 | | Crystal structure of a 2009 H1N1 influenza virus hemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Xu, R, Wilson, I.A. | | 登録日 | 2010-03-01 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus.
Science, 328, 2010
|
|
5HD1
 
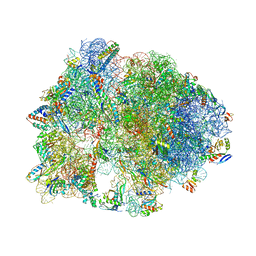 | | Crystal structure of antimicrobial peptide Pyrrhocoricin bound to the Thermus thermophilus 70S ribosome | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | 登録日 | 2016-01-04 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
5YK9
 
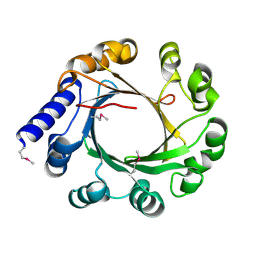 | |
5YDW
 
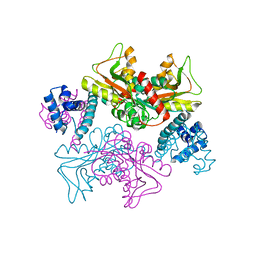 | | Full-length structure of HypT from Salmonella typhimuriuma (hypochlorite-specific LysR-type transcriptional regulator) | | 分子名称: | Cell density-dependent motility repressor | | 著者 | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | 登録日 | 2017-09-15 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1UOT
 
 | | HUMAN CD55 DOMAINS 3 & 4 | | 分子名称: | COMPLEMENT DECAY-ACCELERATING FACTOR | | 著者 | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | 登録日 | 2003-09-23 | | 公開日 | 2003-09-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
3K19
 
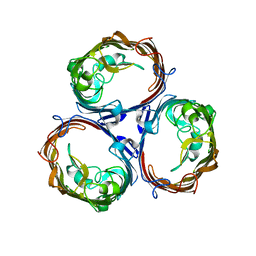 | | OmpF porin | | 分子名称: | Outer membrane protein F | | 著者 | Kefala, G, Ahn, C, Krupa, M, Maslennikov, I, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | 登録日 | 2009-09-26 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.79 Å) | | 主引用文献 | Structures of the OmpF porin crystallized in the presence of foscholine-12.
Protein Sci., 19, 2010
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | 分子名称: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | 著者 | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | 登録日 | 2020-07-14 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
3CMQ
 
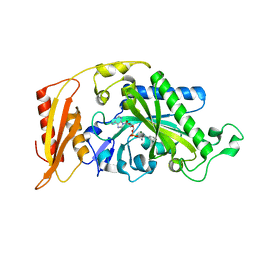 | | Crystal structure of human mitochondrial phenylalanine tRNA synthetase | | 分子名称: | ADENOSINE-5'-[PHENYLALANINYL-PHOSPHATE], MAGNESIUM ION, Phenylalanyl-tRNA synthetase, ... | | 著者 | Klipcan, L, Levin, I.L, Kessler, N, Moor, N, Finarov, I, Safro, M. | | 登録日 | 2008-03-24 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The tRNA-Induced Conformational Activation of Human Mitochondrial Phenylalanyl-tRNA Synthetase.
Structure, 16, 2008
|
|
3I1D
 
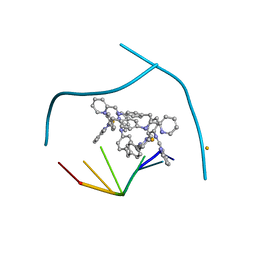 | | Distinct recognition of three-way DNA junctions by the two enantiomers of a metallo-supramolecular cylinder ('helicate') | | 分子名称: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | 著者 | Boer, D.R, Uson, I, Hannon, M.J, Coll, M. | | 登録日 | 2009-06-26 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
4AFL
 
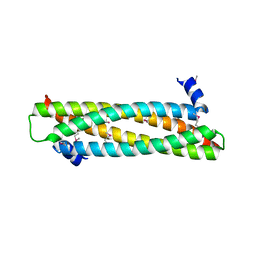 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | 分子名称: | INHIBITOR OF GROWTH PROTEIN 4 | | 著者 | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | 登録日 | 2012-01-19 | | 公開日 | 2012-02-22 | | 最終更新日 | 2012-07-11 | | 実験手法 | X-RAY DIFFRACTION (2.275 Å) | | 主引用文献 | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
5KZ6
 
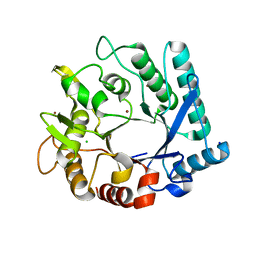 | | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. | | 分子名称: | CHLORIDE ION, Chitinase, SODIUM ION, ... | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-07-22 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.252 Å) | | 主引用文献 | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis.
To Be Published
|
|
3J80
 
 | | CryoEM structure of 40S-eIF1-eIF1A preinitiation complex | | 分子名称: | 18S rRNA, MAGNESIUM ION, RACK1, ... | | 著者 | Hussain, T, Llacer, J.L, Fernandez, I.S, Savva, C.G, Ramakrishnan, V. | | 登録日 | 2014-08-28 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Structural changes enable start codon recognition by the eukaryotic translation initiation complex.
Cell(Cambridge,Mass.), 159, 2014
|
|
